|
EMBOSS
EMBOSS is a free c software analysis package developed for the needs of the molecular biology and bioinformatics user community. The software automatically copes with data in a variety of formats and even allows transparent retrieval of sequence data from the web. Also, as extensive libraries are provided with the package, it is a platform to allow other scientists to develop and release software in true open source spirit. EMBOSS also integrates a range of currently available packages and tools for sequence analysis into a seamless whole. ''EMBOSS'' is an acronym for European Molecular Biology Open Software Suite. The ''European'' part of the name hints at the wider scope. The core EMBOSS groups are collaborating with many other groups to develop the new applications that the users need. This was done from the beginning with EMBnet, the European Molecular Biology Network. EMBnet has many nodes worldwide most of which are national bioinformatics services. EMBnet has the programmi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EMBnet
The European Molecular Biology network (EMBnet) is an international scientific network and interest group that aims to enhance bioinformatics services by bringing together bioinformatics expertises and capacities. On 2011 EMBnet has 37 nodes spread over 32 countries. The nodes include bioinformatics related university departments, research institutes and national service providers. Operations The main task of most EMBnet nodes is to provide their national scientific community with access to bioinformatics databanks, specialised software and sufficient computing resources and expertise. EMBnet is also working in the fields of bioinformatics training and software development. Examples of software created by EMBnet members are: EMBOSS, wEMBOSS, UTOPIA. EMBnet represents a wide user group and works closely together with the database producers such as EMBL's European Bioinformatics Institute (EBI), the Swiss Institute of Bioinformatics (Swiss-Prot), the Munich Information Center for P ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural biology, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are typically represented as rows within a matrix (mathematics), matrix. Gaps are inserted between the Residue (chemistry), residues so that identical or similar characters are aligned in successive columns. Sequence alignments are also used for non-biological sequences such as calculating the Edit distance, distance cost between strings in a natural language, or to display financial data. Interpretation If two sequences in an alignment share a common ancestor, mismatches can be interpreted as point mutations and gaps as indels (that is, insertion or deletion mutations) introduced in one or both lineages in the time since they diverged from one another. In sequence ali ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Open Bioinformatics Foundation
The Open Bioinformatics Foundation is a non-profit, volunteer-run organization focused on supporting open source programming in bioinformatics. The mission of the foundation is to support the development of open source toolkits for bioinformatics, organise developer-centric hackathon events and generally assist in the development and promotion of open source software development in the life sciences. The foundation also organises and runs the annual Bioinformatics Open Source Conference, a satellite meeting of the Intelligent Systems for Molecular Biology conference. The foundation participates in the Google Summer of Code, acting as an umbrella organisation for individual bioinformatics-related projects. The Open Bioinformatics Foundation was started in 2001, arising from the BioJava, BioPerl and BioPython projects. A formal membership for the foundation was created in 2005. In October 2012, the foundation began an association with Software in the Public Interest (SPI), a US ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatics
Bioinformatics () is an interdisciplinary field of science that develops methods and Bioinformatics software, software tools for understanding biological data, especially when the data sets are large and complex. Bioinformatics uses biology, chemistry, physics, computer science, data science, computer programming, information engineering, mathematics and statistics to analyze and interpret biological data. The process of analyzing and interpreting data can sometimes be referred to as computational biology, however this distinction between the two terms is often disputed. To some, the term ''computational biology'' refers to building and using models of biological systems. Computational, statistical, and computer programming techniques have been used for In silico, computer simulation analyses of biological queries. They include reused specific analysis "pipelines", particularly in the field of genomics, such as by the identification of genes and single nucleotide polymorphis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genostar
Genostar is a bioinformatics provider based in Grenoble, France. The company was founded in 2004 following the "Genostar consortium" that was created in 1999 as a public-private consortium by Genome Express, Hybrigenics, INRIA (Institut National de Recherche en Informatique et Automatique / French National Institute for Research in Computer Science and Control) and The Pasteur Institute. Software Metabolic Pathway Builder is a bioinformatics environment dedicated to microbial research. This covers sequence assembly, mapping, annotation transfer and identification of protein domains, comparative genomics, structural searches, metabolic pathway analysis, modeling and simulation of biological networks. Genostar's software is platform independent and can thus be used for both Mac OS X, Windows, and Linux. Sequence assembly *Mapping an ensemble of sequences on a reference sequence *between a reference sequence and contigs, between two sequences or between two sets of sequences *findi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
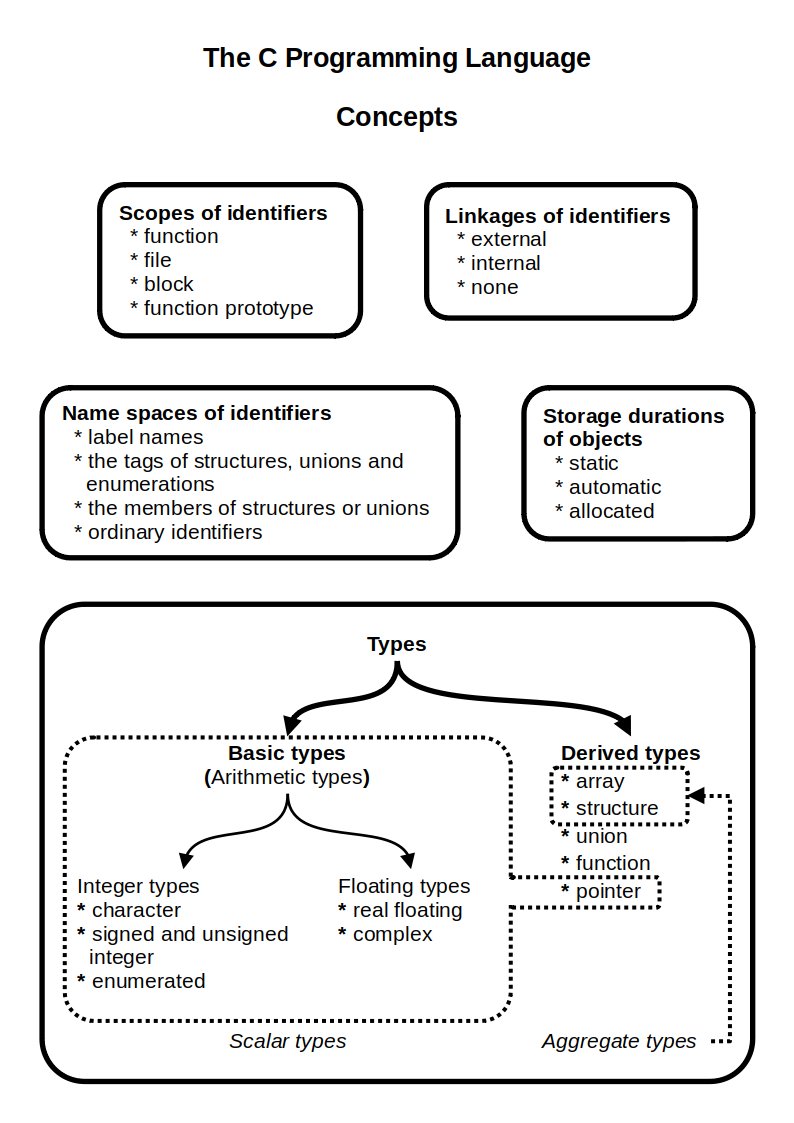
C (programming Language)
C (''pronounced'' '' – like the letter c'') is a general-purpose programming language. It was created in the 1970s by Dennis Ritchie and remains very widely used and influential. By design, C's features cleanly reflect the capabilities of the targeted Central processing unit, CPUs. It has found lasting use in operating systems code (especially in Kernel (operating system), kernels), device drivers, and protocol stacks, but its use in application software has been decreasing. C is commonly used on computer architectures that range from the largest supercomputers to the smallest microcontrollers and embedded systems. A successor to the programming language B (programming language), B, C was originally developed at Bell Labs by Ritchie between 1972 and 1973 to construct utilities running on Unix. It was applied to re-implementing the kernel of the Unix operating system. During the 1980s, C gradually gained popularity. It has become one of the most widely used programming langu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Science And Technology In Cambridgeshire
Science is a systematic discipline that builds and organises knowledge in the form of testable hypotheses and predictions about the universe. Modern science is typically divided into twoor threemajor branches: the natural sciences, which study the physical world, and the social sciences, which study individuals and societies. While referred to as the formal sciences, the study of logic, mathematics, and theoretical computer science are typically regarded as separate because they rely on deductive reasoning instead of the scientific method as their main methodology. Meanwhile, applied sciences are disciplines that use scientific knowledge for practical purposes, such as engineering and medicine. The history of science spans the majority of the historical record, with the earliest identifiable predecessors to modern science dating to the Bronze Age in Egypt and Mesopotamia (). Their contributions to mathematics, astronomy, and medicine entered and shaped the Greek natural philo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Free Science Software
Free may refer to: Concept * Freedom, the ability to act or change without constraint or restriction * Emancipate, attaining civil and political rights or equality * Free (''gratis''), free of charge * Gratis versus libre, the difference between the two common meanings of the adjective "free". Computing * Free (programming), a function that releases dynamically allocated memory for reuse * Free software, software usable and distributable with few restrictions and no payment *, an emoji in the Enclosed Alphanumeric Supplement block. Mathematics * Free object ** Free abelian group ** Free algebra ** Free group ** Free module ** Free semigroup * Free variable People * Free (surname) * Free (rapper) (born 1968), or Free Marie, American rapper and media personality * Free, a pseudonym for the activist and writer Abbie Hoffman * Free (active 2003–), American musician in the band FreeSol Arts and media Film and television * ''Free'' (film), a 2001 American dramed ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatics Software
{{short description, none The list of bioinformatics Bioinformatics () is an interdisciplinary field of science that develops methods and Bioinformatics software, software tools for understanding biological data, especially when the data sets are large and complex. Bioinformatics uses biology, ... software tools can be split up according to the license used: * List of proprietary bioinformatics software * List of open-source bioinformatics software Alternatively, here is a categorization according to the respective bioinformatics subfield specialized on: *Sequence analysis software ** List of sequence alignment software ** List of alignment visualization software ** Alignment-free sequence analysis ** De novo sequence assemblers ** List of gene prediction software ** List of disorder prediction software ** List of Protein subcellular localization prediction tools ** List of phylogenetics software ** List of phylogenetic tree visualization software ** :Metagenomics_software *S ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SOAP (protocol)
Soap is a salt of a fatty acid (sometimes other carboxylic acids) used for cleaning and lubricating products as well as other applications. In a domestic setting, soaps, specifically "toilet soaps", are surfactants usually used for washing, bathing, and other types of housekeeping. In industrial settings, soaps are used as thickeners, components of some lubricants, emulsifiers, and catalysts. Soaps are often produced by mixing fats and oils with a base. Humans have used soap for millennia; evidence exists for the production of soap-like materials in ancient Babylon around 2800 BC. Types Toilet soaps In a domestic setting, "soap" usually refers to what is technically called a toilet soap, used for household and personal cleaning. Toilet soaps are salts of fatty acids with the general formula ( RCO2−)M+, where M is Na (sodium) or K (potassium). When used for cleaning, soap solubilizes particles and grime, which can then be separated from the article being cleaned. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GNU Library General Public Licence
The GNU Lesser General Public License (LGPL) is a free-software license published by the Free Software Foundation (FSF). The license allows developers and companies to use and integrate a software component released under the LGPL into their own (even proprietary) software without being required by the terms of a strong copyleft license to release the source code of their own components. However, any developer who modifies an LGPL-covered component is required to make their modified version available under the same LGPL license. For proprietary software, code under the LGPL is usually used in the form of a shared library, so that there is a clear separation between the proprietary and LGPL components. The LGPL is primarily used for software libraries, although it is also used by some stand-alone applications. The LGPL was developed as a compromise between the strong copyleft of the GNU General Public License (GPL) and more permissive licenses such as the BSD licenses and the MIT L ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |