|
Duplex Sequencing
Duplex sequencing is a library preparation and analysis method for next-generation sequencing (NGS) platforms that employs random tagging of double-stranded DNA to detect mutations with higher accuracy and lower error rates. This method uses degenerate molecular tags in addition to sequencing adapters to recognize reads originating from each strand of DNA. The generated sequencing reads then will be analyzed using two methods: single-strand consensus sequences (SSCS) and duplex consensus sequences (DCS) assembly. Duplex sequencing theoretically can detect mutations with frequencies as low as 5 x 10−8 --that is more than 10,000 times higher in accuracy compared to the conventional next-generation sequencing methods.M. W. Schmitt, S. R. Kennedy, J. J. Salk, et al“Detection of ultra-rare mutations by next-generation sequencing” Proc. Natl. Acad. Sci., vol. 109 no. 36. 2012. .S. R. Kennedy, M. W. Schmitt, E. J. Fox, B. F. Kohrn, et al Nature Protoc., vol. 9 no. 11, 2586-606. 2 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Duplex Sequencing Overview Alphabeta Fix
Duplex (Latin, 'double') may refer to: Arts and entertainment * ''Duplex'' (film), or ''Our House'', a 2003 American black comedy film * Duplex (band), a Dutch electronic music duo * Duplex (Norwegian duo) * Duplex!, a Canadian children's music band * '' The Duplex'', an American comic strip by Glenn McCoy * ''The Duplex'' (film), a 2015 Nigerian film Places * Duplex, Tennessee, U.S. * Duplex, Texas, U.S. Science and technology * ''Duplex'' (moth), a genus of moths in the family Erebidae * Nucleic acid double helix, a double-stranded molecule of DNA or RNA * Duplex (telecommunications), communications in both directions simultaneously * Duplex (typography), a Linotype technique * Duplex ultrasonography, a medical imaging technique * Google Duplex, an extension of Google Assistant Transportation * Duplex (automobile), an early car built in Canada in 1923 * Duplex locomotive, a type of steam locomotive * TGV Duplex, a French high-speed train of the TGV family featuring bi-le ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ligation (molecular Biology)
In molecular biology, ligation is the joining of two nucleic acid fragments through the action of an enzyme. It is an essential laboratory procedure in the molecular cloning of DNA, whereby DNA fragments are joined to create recombinant DNA molecules (such as when a foreign DNA fragment is inserted into a plasmid). The ends of DNA fragments are joined by the formation of phosphodiester bonds between the 3'-hydroxyl of one DNA terminus with the 5'-phosphoryl of another. RNA may also be ligated similarly. A co-factor is generally involved in the reaction, and this is usually ATP or NAD+. Ligation in the laboratory is normally performed using T4 DNA ligase. However, procedures for ligation without the use of standard DNA ligase are also popular. Ligation reaction The mechanism of the ligation reaction was first elucidated in the laboratory of I. Robert Lehman. Two fragments of DNA may be joined by DNA ligase which catalyzes the formation of a phosphodiester bond between the 3' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cancer Genome Sequencing
Cancer genome sequencing is the whole genome sequencing of a single, homogeneous or heterogeneous group of cancer cells. It is a biochemical laboratory method for the characterization and identification of the DNA or RNA sequences of cancer cell(s). Unlike whole genome (WG) sequencing which is typically from blood cells, such as J. Craig Venter's and James D. Watson’s WG sequencing projects, saliva, epithelial cells or bone - cancer genome sequencing involves direct sequencing of primary tumor tissue, adjacent or distal normal tissue, the tumor micro environment such as fibroblast/stromal cells, or metastatic tumor sites. Similar to whole genome sequencing, the information generated from this technique include: identification of nucleotide bases (DNA or RNA), copy number and sequence variants, mutation status, and structural changes such as chromosomal translocations and fusion genes. Cancer genome sequencing is not limited to WG sequencing and can also include exome, transcrip ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
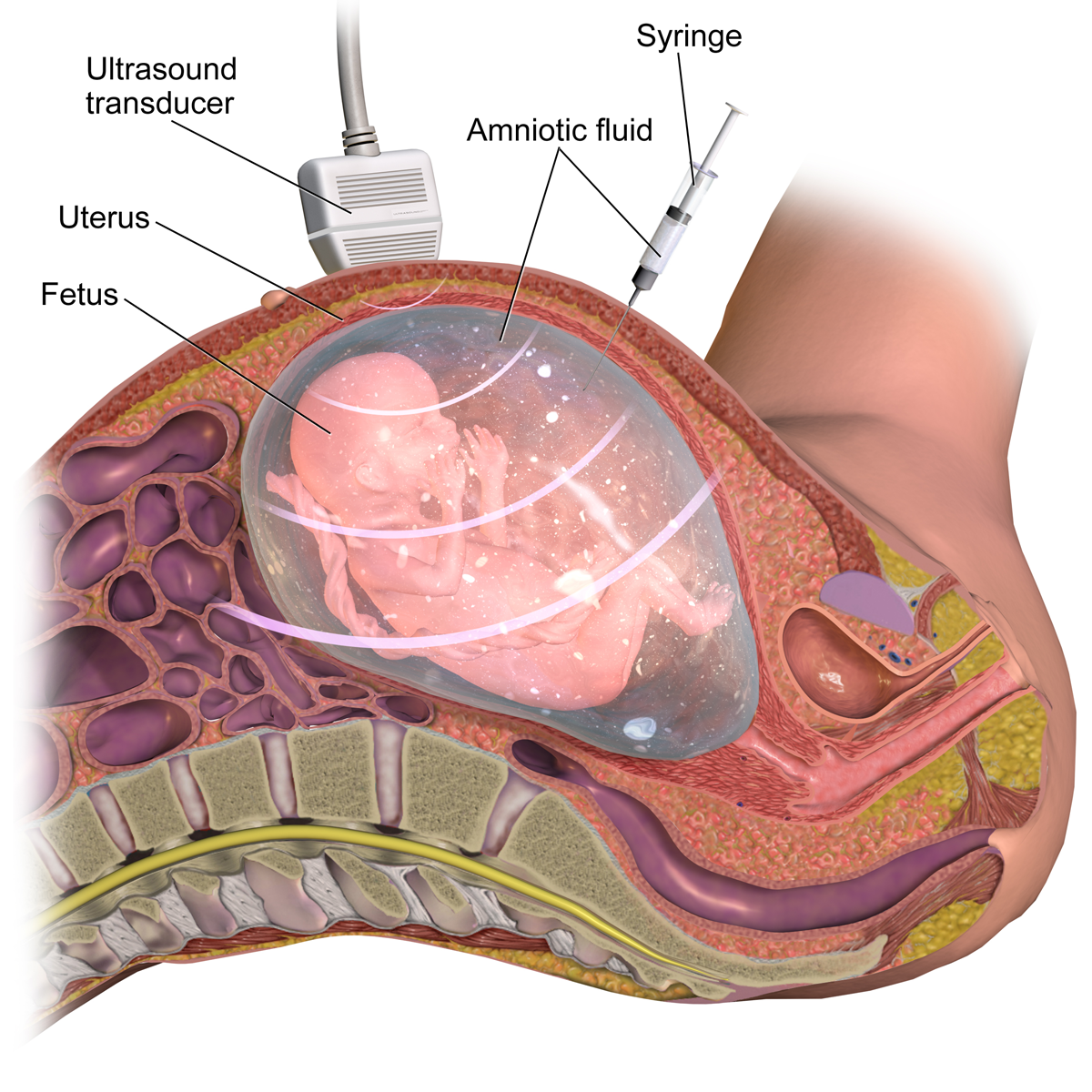
Prenatal Diagnosis
Prenatal testing consists of prenatal screening and prenatal diagnosis, which are aspects of prenatal care that focus on detecting problems with the pregnancy as early as possible. These may be anatomic and physiologic problems with the health of the zygote, embryo, or fetus, either before gestation even starts (as in preimplantation genetic diagnosis) or as early in gestation as practicable. Screening can detect problems such as neural tube defects, chromosome abnormalities, and gene mutations that would lead to genetic disorders and birth defects, such as spina bifida, cleft palate, Down syndrome, Tay–Sachs disease, sickle cell anemia, thalassemia, cystic fibrosis, muscular dystrophy, and fragile X syndrome. Some tests are designed to discover problems which primarily affect the health of the mother, such as PAPP-A to detect pre-eclampsia or glucose tolerance tests to diagnose gestational diabetes. Screening can also detect anatomical defects such as hydrocephalus, anenceph ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
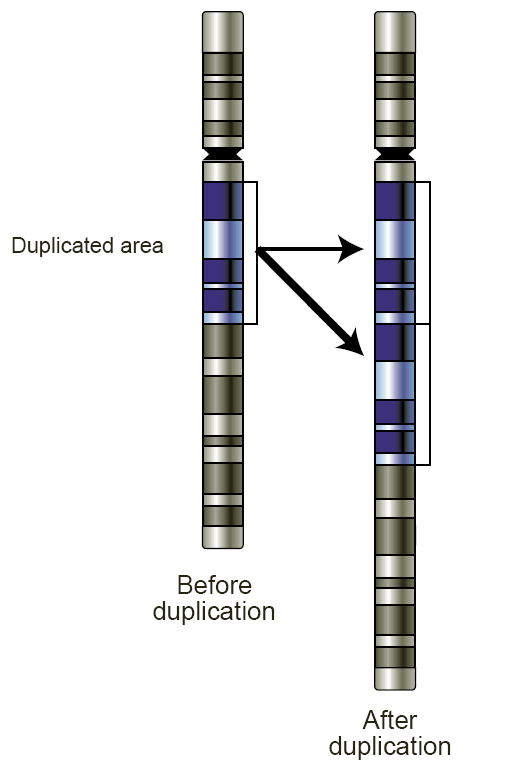
Copy-number Variation
Copy number variation (CNV) is a phenomenon in which sections of the genome are repeated and the number of repeats in the genome varies between individuals. Copy number variation is a type of structural variation: specifically, it is a type of duplication or deletion event that affects a considerable number of base pairs. Approximately two-thirds of the entire human genome may be composed of repeats and 4.8–9.5% of the human genome can be classified as copy number variations. In mammals, copy number variations play an important role in generating necessary variation in the population as well as disease phenotype. Copy number variations can be generally categorized into two main groups: short repeats and long repeats. However, there are no clear boundaries between the two groups and the classification depends on the nature of the loci of interest. Short repeats include mainly dinucleotide repeats (two repeating nucleotides e.g. A-C-A-C-A-C...) and trinucleotide repeats. Long r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
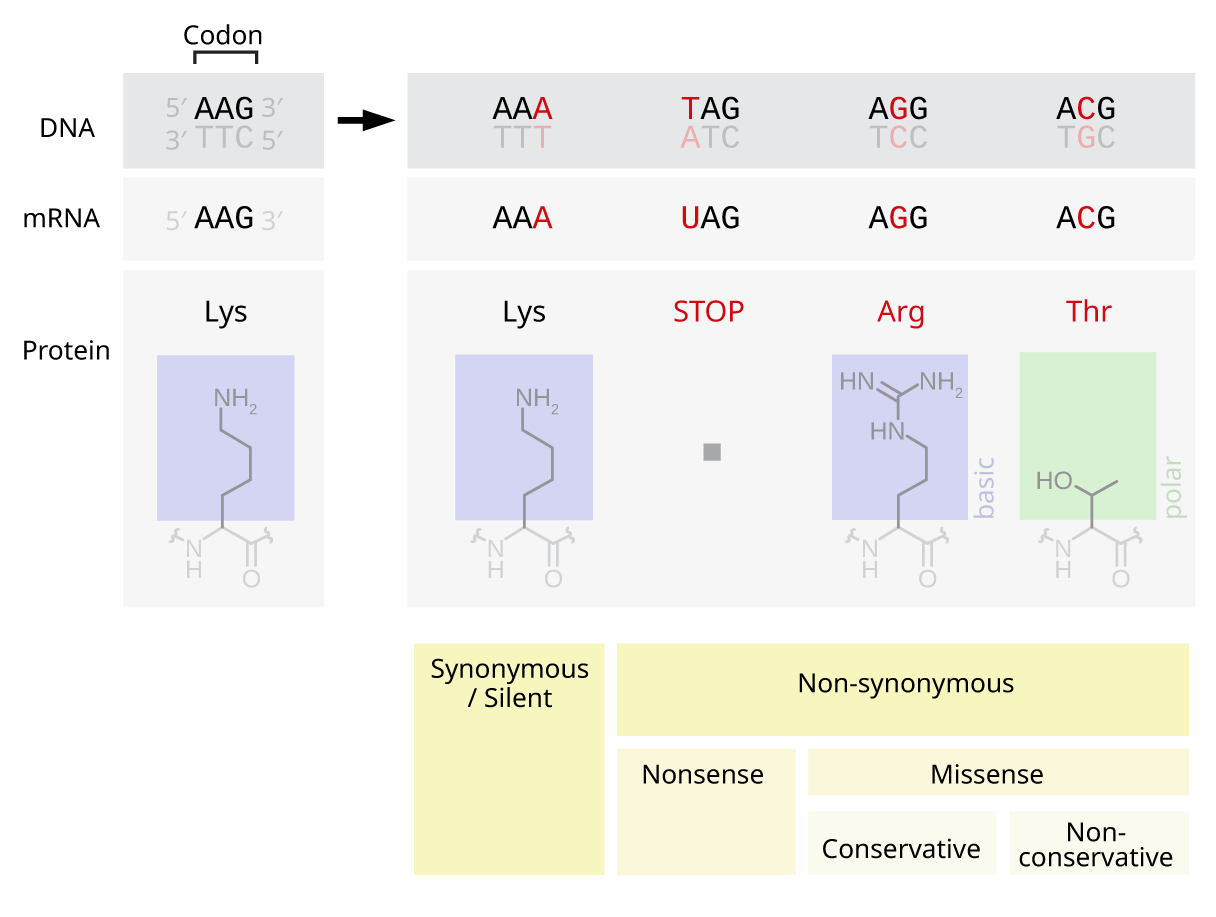
Point Mutation
A point mutation is a genetic mutation where a single nucleotide base is changed, inserted or deleted from a DNA or RNA sequence of an organism's genome. Point mutations have a variety of effects on the downstream protein product—consequences that are moderately predictable based upon the specifics of the mutation. These consequences can range from no effect (e.g. synonymous mutations) to deleterious effects (e.g. frameshift mutations), with regard to protein production, composition, and function. Causes Point mutations usually take place during DNA replication. DNA replication occurs when one double-stranded DNA molecule creates two single strands of DNA, each of which is a template for the creation of the complementary strand. A single point mutation can change the whole DNA sequence. Changing one purine or pyrimidine may change the amino acid that the nucleotides code for. Point mutations may arise from spontaneous mutations that occur during DNA replication. The rate of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transversion
Transversion, in molecular biology, refers to a point mutation in DNA in which a single (two ring) purine ( A or G) is changed for a (one ring) pyrimidine ( T or C), or vice versa. A transversion can be spontaneous, or it can be caused by ionizing radiation or alkylating agents. It can only be reversed by a spontaneous reversion. Ratio of transitions to transversions Although there are two possible transversions but only one possible transition per base, transition mutations are more likely than transversions because substituting a single ring structure for another single ring structure is more likely than substituting a double ring for a single ring. Also, transitions are less likely to result in amino acid substitutions (due to wobble base pair), and are therefore more likely to persist as "silent substitutions" in populations as single nucleotide polymorphisms (SNPs). A transversion usually has a more pronounced effect than a transition because the third nucleotide c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Sequence Alignment Software
This list of sequence alignment software is a compilation of software tools and web portals used in pairwise sequence alignment and multiple sequence alignment. See structural alignment software for structural alignment of proteins. Database search only *Sequence type: protein or nucleotide Pairwise alignment *Sequence type: protein or nucleotide **Alignment type: local or global Multiple sequence alignment *Sequence type: protein or nucleotide. **Alignment type: local or global Genomics analysis *Sequence type: protein or nucleotide Motif finding *Sequence type: protein or nucleotide Benchmarking Alignment viewers, editors Please see List of alignment visualization software. Short-read sequence alignment See also * List of open source bioinformatics software References {{Reflist Genetics-related lists, Sequence Lists of bioinformatics software, Sequence alignment software ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reference Genome
A reference genome (also known as a reference assembly) is a digital nucleic acid sequence database, assembled by scientists as a representative example of the set of genes in one idealized individual organism of a species. As they are assembled from the sequencing of DNA from a number of individual donors, reference genomes do not accurately represent the set of genes of any single individual organism. Instead a reference provides a haploid mosaic of different DNA sequences from each donor. For example, the most recent human reference genome (assembly '' GRCh38/hg38'') is derived from >60 genomic clone libraries. There are reference genomes for multiple species of viruses, bacteria, fungus, plants, and animals. Reference genomes are typically used as a guide on which new genomes are built, enabling them to be assembled much more quickly and cheaply than the initial Human Genome Project. Reference genomes can be accessed online at several locations, using dedicated browsers ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Aligned sequences of nucleotide or amino acid residues are typically represented as rows within a matrix. Gaps are inserted between the residues so that identical or similar characters are aligned in successive columns. Sequence alignments are also used for non-biological sequences, such as calculating the distance cost between strings in a natural language or in financial data. Interpretation If two sequences in an alignment share a common ancestor, mismatches can be interpreted as point mutations and gaps as indels (that is, insertion or deletion mutations) introduced in one or both lineages in the time since they diverged from one another. In sequence alignments of proteins, the degree of similarity between amino acids occupying a pa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Consensus Sequence
In molecular biology and bioinformatics, the consensus sequence (or canonical sequence) is the calculated order of most frequent residues, either nucleotide or amino acid, found at each position in a sequence alignment. It serves as a simplified representation of the population. It represents the results of multiple sequence alignments in which related sequences are compared to each other and similar sequence motifs are calculated. Such information is important when considering sequence-dependent enzymes such as RNA polymerase.Pierce, Benjamin A. 2002. Genetics : A Conceptual Approach. 1st ed. New York: W.H. Freeman and Co. Biological significance A protein binding site, represented by a consensus sequence, may be a short sequence of nucleotides which is found several times in the genome and is thought to play the same role in its different locations. For example, many transcription factors recognize particular patterns in the promoters of the genes they regulate. In the same way ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Upstream And Downstream (DNA)
In molecular biology and genetics, upstream and downstream both refer to relative positions of genetic code in DNA or RNA. Each strand of DNA or RNA has a 5' end and a 3' end, so named for the carbon position on the deoxyribose Deoxyribose, or more precisely 2-deoxyribose, is a monosaccharide with idealized formula H−(C=O)−(CH2)−(CHOH)3−H. Its name indicates that it is a deoxy sugar, meaning that it is derived from the sugar ribose by loss of a hydroxy group ... (or ribose) ring. By convention, upstream and downstream relate to the 5' to 3' direction respectively in which RNA transcription takes place. Upstream is toward the 5' end of the RNA molecule and downstream is toward the 3' end. When considering double-stranded DNA, upstream is toward the 5' end of the coding strand for the gene in question and downstream is toward the 3' end. Due to the anti-parallel nature of DNA, this means the 3' end of the template strand is upstream of the gene and the 5' end is d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |