|
Chou–Fasman Method
The Chou–Fasman method is an empirical technique for the prediction of secondary structures in proteins, originally developed in the 1970s by Peter Y. Chou and Gerald D. Fasman. The method is based on analyses of the relative frequencies of each amino acid in alpha helices, beta sheets, and turns based on known protein structures solved with X-ray crystallography. From these frequencies a set of probability parameters were derived for the appearance of each amino acid in each secondary structure type, and these parameters are used to predict the probability that a given sequence of amino acids would form a helix, a beta strand, or a turn in a protein. The method is at most about 50–60% accurate in identifying correct secondary structures, which is significantly less accurate than the modern machine learning–based techniques. Amino acid propensities The original Chou–Fasman parameters found some strong tendencies among individual amino acids to prefer one type of secondary s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Protein Structure Prediction
Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequence—that is, the prediction of its Protein secondary structure, secondary and Protein tertiary structure, tertiary structure from Protein primary structure, primary structure. Structure prediction is different from the inverse problem of protein design. Protein structure prediction is one of the most important goals pursued by computational biology and addresses Levinthal's paradox. Accurate structure prediction has important applications in medicine (for example, in drug design) and biotechnology (for example, in novel enzyme design). Starting in 1994, the performance of current methods is assessed biannually in the ''Critical Assessment of Structure Prediction'' (CASP) experiment. A continuous evaluation of protein structure prediction web servers is performed by the community project ''Continuous Automated Model EvaluatiOn'' (CAMEO3D). Protein structure a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Alanine
Alanine (symbol Ala or A), or α-alanine, is an α-amino acid that is used in the biosynthesis of proteins. It contains an amine group and a carboxylic acid group, both attached to the central carbon atom which also carries a methyl group side chain. Consequently it is classified as a non-polar, aliphatic α-amino acid. Under biological conditions, it exists in its zwitterionic form with its amine group protonated (as ) and its carboxyl group deprotonated (as ). It is non-essential to humans as it can be synthesized metabolically and does not need to be present in the diet. It is encoded by all codons starting with G C (GC U, GCC, GC A, and GCG). The L-isomer of alanine (left-handed) is the one that is incorporated into proteins. L-alanine is second only to L-leucine in rate of occurrence, accounting for 7.8% of the primary structure in a sample of 1,150 proteins. The right-handed form, D-alanine, occurs in peptides in some bacterial cell walls (in peptidoglycan) ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
List Of Protein Structure Prediction Software
This list of protein structure prediction software summarizes notable used software tools in protein structure prediction, including homology modeling, protein threading, ''ab initio'' methods, secondary structure prediction, and transmembrane helix and signal peptide prediction. Software list Below is a list which separates programs according to the method used for structure prediction. Homology modeling Threading and fold recognition ''Ab initio'' structure prediction Secondary structure prediction Detailed list of programs can be found at List of protein secondary structure prediction programs See also * List of protein secondary structure prediction programs *Comparison of nucleic acid simulation software *List of software for molecular mechanics modeling *Molecular design software * Protein design *AlphaFold AlphaFold is an artificial intelligence (AI) program developed by DeepMind, a subsidiary of Alphabet, which performs predictions of protein struc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
GOR Method
The GOR method (short for Garnier–Osguthorpe–Robson) is an information theory-based method for the prediction of secondary structures in proteins. It was developed in the late 1970s shortly after the simpler Chou–Fasman method. Like Chou–Fasman, the GOR method is based on probability parameters derived from empirical studies of known protein tertiary structures solved by X-ray crystallography. However, unlike Chou–Fasman, the GOR method takes into account not only the propensities of individual amino acids to form particular secondary structures, but also the conditional probability of the amino acid to form a secondary structure given that its immediate neighbors have already formed that structure. The method is therefore essentially Bayesian in its analysis. Method The GOR method analyzes sequences to predict alpha helix, beta sheet, turn, or random coil secondary structure at each position based on 17-amino-acid sequence windows. The original description of the method i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
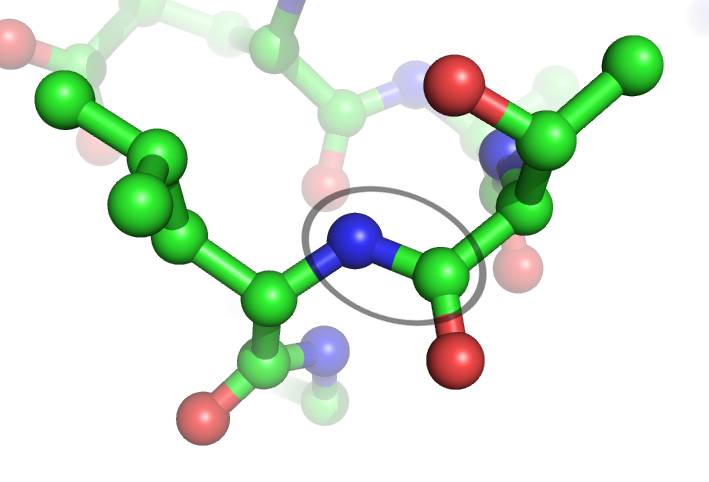
Peptide Bond
In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 (carbon number one) of one alpha-amino acid and N2 (nitrogen number two) of another, along a peptide or protein chain. It can also be called a eupeptide bond to distinguish it from an isopeptide bond, which is another type of amide bond between two amino acids. Synthesis When two amino acids form a '' dipeptide'' through a ''peptide bond'', it is a type of condensation reaction. In this kind of condensation, two amino acids approach each other, with the non-side chain (C1) carboxylic acid moiety of one coming near the non-side chain (N2) amino moiety of the other. One loses a hydrogen and oxygen from its carboxyl group (COOH) and the other loses a hydrogen from its amino group (NH2). This reaction produces a molecule of water (H2O) and two amino acids joined by a peptide bond (−CO−NH−). The two joined amino acids are called a dipeptide. The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Glycine
Glycine (symbol Gly or G; ) is an amino acid that has a single hydrogen atom as its side chain. It is the simplest stable amino acid. Glycine is one of the proteinogenic amino acids. It is encoded by all the codons starting with GG (GGU, GGC, GGA, GGG). Glycine disrupts the formation of alpha-helices in secondary protein structure. Its small side chain causes it to favor random coils instead. Glycine is also an inhibitory neurotransmitter – interference with its release within the spinal cord (such as during a '' Clostridium tetani'' infection) can cause spastic paralysis due to uninhibited muscle contraction. It is the only achiral proteinogenic amino acid. It can fit into both hydrophilic and hydrophobic environments, due to its minimal side chain of only one hydrogen atom. History and etymology Glycine was discovered in 1820 by French chemist Henri Braconnot when he hydrolyzed gelatin by boiling it with sulfuric acid. He originally called it "sugar of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Proline
Proline (symbol Pro or P) is an organic acid classed as a proteinogenic amino acid (used in the biosynthesis of proteins), although it does not contain the amino group but is rather a secondary amine. The secondary amine nitrogen is in the protonated form (NH2+) under biological conditions, while the carboxyl group is in the deprotonated −COO− form. The "side chain" from the α carbon connects to the nitrogen forming a pyrrolidine loop, classifying it as a aliphatic amino acid. It is non-essential in humans, meaning the body can synthesize it from the non-essential amino acid L-glutamate. It is encoded by all the codons starting with CC (CCU, CCC, CCA, and CCG). Proline is the only proteinogenic amino acid which is a secondary amine, as the nitrogen atom is attached both to the α-carbon and to a chain of three carbons that together form a five-membered ring. History and etymology Proline was first isolated in 1900 by Richard Willstätter who obtained the amino a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Methionine
Methionine (symbol Met or M) () is an essential amino acid in humans. As the precursor of other non-essential amino acids such as cysteine and taurine, versatile compounds such as SAM-e, and the important antioxidant glutathione, methionine plays a critical role in the metabolism and health of many species, including humans. Methionine is also involved in angiogenesis and various processes related to DNA transcription, epigenetic expression, and gene regulation. Methionine was first isolated in 1921 by John Howard Mueller. It is Genetic code, encoded by the codon AUG. It was named by Satoru Odake in 1925, as an abbreviation of its structural description 2-amino-4-(methylthio)butanoic acid. Biochemical details Methionine (abbreviated as Met or M; encoded by the codon AUG) is an α-amino acid that is used in the biosynthesis of proteins. It contains a carboxyl group (which is in the deprotonated −COO− form under biological pH conditions), an amino group (which is in the proton ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Leucine
Leucine (symbol Leu or L) is an essential amino acid that is used in the biosynthesis of proteins. Leucine is an α-amino acid, meaning it contains an α-amino group (which is in the protonated −NH3+ form under biological conditions), an α-Carboxylic acid, carboxylic acid group (which is in the deprotonated −COO− form under biological conditions), and a side chain Isobutyl, isobutyl group, making it a Chemical polarity, non-polar Aliphatic compound, aliphatic amino acid. It is Essential amino acid, essential in humans, meaning the body cannot synthesize it; it must be obtained from the diet. Human dietary sources are foods that contain protein, such as meats, dairy products, soy products, and beans and other legumes. It is genetic code, encoded by the codons UUA, UUG, CUU, CUC, CUA, and CUG. Leucine is named after the Greek language, Greek word for "white": ''λευκός'' (''leukós'', "white"), after its common appearance as a white powder, a property it shares with many ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Glutamate
Glutamic acid (symbol Glu or E; known as glutamate in its anionic form) is an α-amino acid that is used by almost all living beings in the biosynthesis of proteins. It is a Essential amino acid, non-essential nutrient for humans, meaning that the human body can synthesize enough for its use. It is also the most abundant excitatory neurotransmitter in the vertebrate nervous system. It serves as the precursor for the synthesis of the inhibitory gamma-aminobutyric acid (GABA) in GABAergic neurons. Its molecular formula is . Glutamic acid exists in two optically isomeric forms; the optical rotation, dextrorotatory -form is usually obtained by hydrolysis of gluten or from the waste waters of beet-sugar manufacture or by fermentation.Webster's Third New International Dictionary of the English Language Unabridged, Third Edition, 1971. Its molecular structure could be idealized as HOOC−CH()−()2−COOH, with two carboxylic acid, carboxyl groups −COOH and one amine, amino group � ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Machine Learning
Machine learning (ML) is a field of study in artificial intelligence concerned with the development and study of Computational statistics, statistical algorithms that can learn from data and generalise to unseen data, and thus perform Task (computing), tasks without explicit Machine code, instructions. Within a subdiscipline in machine learning, advances in the field of deep learning have allowed Neural network (machine learning), neural networks, a class of statistical algorithms, to surpass many previous machine learning approaches in performance. ML finds application in many fields, including natural language processing, computer vision, speech recognition, email filtering, agriculture, and medicine. The application of ML to business problems is known as predictive analytics. Statistics and mathematical optimisation (mathematical programming) methods comprise the foundations of machine learning. Data mining is a related field of study, focusing on exploratory data analysi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Secondary Structure
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |