|
Cyclophilin A
Peptidylprolyl isomerase A (PPIA), also known as cyclophilin A (CypA) or rotamase A is an enzyme that in humans is encoded by the ''PPIA'' gene on chromosome 7. As a member of the peptidyl-prolyl cis-trans isomerase (PPIase) family, this protein catalyzes the cis-trans isomerization of proline imidic peptide bonds, which allows it to regulate many biological processes, including signal transduction, intracellular signaling, transcription (genetics), transcription, inflammation, and apoptosis. Due to its various functions, PPIA has been implicated in a broad range of inflammatory diseases, including atherosclerosis and arthritis, and viral infections. Structure PPIA is an 18 kDa, 165-amino acid long cytosolic protein. Like other cyclophilins, PPIA forms a β-barrel structure with a hydrophobic core. This β-barrel is composed of eight anti-parallel β-strands and capped by two α-helices at the top and bottom. In addition, the β-turns and loops in the strands contribute to the fle ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PPIase
Prolyl isomerase (also known as peptidylprolyl isomerase or PPIase) is an enzyme () found in both prokaryotes and eukaryotes that interconverts the ''cis'' and ''trans'' isomers of peptide bonds with the amino acid proline. Proline has an unusually conformationally restrained peptide bond due to its cyclic structure with its side chain bonded to its secondary amine nitrogen. Most amino acids have a strong energetic preference for the ''trans'' peptide bond conformation due to steric hindrance, but proline's unusual structure stabilizes the ''cis'' form so that both isomers are populated under biologically relevant conditions. Proteins with prolyl isomerase activity include cyclophilin, FKBPs, and parvulin, although larger proteins can also contain prolyl isomerase structural domain, domains. Protein folding Proline is unique among the natural amino acids in having a relatively small difference in free energy between the ''cis'' configuration of its peptide bond and the more c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NF-κB
Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) is a protein complex that controls transcription of DNA, cytokine production and cell survival. NF-κB is found in almost all animal cell types and is involved in cellular responses to stimuli such as stress, cytokines, free radicals, heavy metals, ultraviolet irradiation, oxidized LDL, and bacterial or viral antigens. NF-κB plays a key role in regulating the immune response to infection. Incorrect regulation of NF-κB has been linked to cancer, inflammatory and autoimmune diseases, septic shock, viral infection, and improper immune development. NF-κB has also been implicated in processes of synaptic plasticity and memory. Discovery NF-κB was discovered by Ranjan Sen in the lab of Nobel laureate David Baltimore via its interaction with an 11-base pair sequence in the immunoglobulin light-chain enhancer in B cells. Later work by Alexander Poltorak and Bruno Lemaitre in mice and ''Drosophila'' frui ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reactive Oxygen Species
In chemistry, reactive oxygen species (ROS) are highly reactive chemicals formed from diatomic oxygen (). Examples of ROS include peroxides, superoxide, hydroxyl radical, singlet oxygen, and alpha-oxygen. The reduction of molecular oxygen () produces superoxide (), which is the precursor to most other reactive oxygen species: :O2 + e^- -> \ ^\bullet O2- Dismutation of superoxide produces hydrogen peroxide (): :2 H+ + \ ^\bullet O2^- + \ ^\bullet O2^- -> H2O2 + O2 Hydrogen peroxide in turn may be partially reduced, thus forming hydroxide ions and hydroxyl radicals (), or fully reduced to water: :H2O2 + e^- -> HO^- + \ ^\bullet OH :2 H+ + 2 e- + H2O2 -> 2 H2O In a biological context, ROS are byproducts of the normal metabolism of oxygen. ROS have roles in cell signaling and homeostasis. ROS are intrinsic to cellular functioning, and are present at low and stationary levels in normal cells. In plants, ROS are involved in metabolic processes related to photoprotection and toleran ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitochondria
A mitochondrion (; ) is an organelle found in the Cell (biology), cells of most Eukaryotes, such as animals, plants and Fungus, fungi. Mitochondria have a double lipid bilayer, membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used throughout the cell as a source of chemical energy. They were discovered by Albert von Kölliker in 1857 in the voluntary muscles of insects. The term ''mitochondrion'' was coined by Carl Benda in 1898. The mitochondrion is popularly nicknamed the "powerhouse of the cell", a phrase coined by Philip Siekevitz in a 1957 article of the same name. Some cells in some multicellular organisms lack mitochondria (for example, mature mammalian red blood cells). A large number of unicellular organisms, such as microsporidia, parabasalids and diplomonads, have reduced or transformed their mitochondria into mitosome, other structures. One eukaryote, ''Monocercomonoides'', is known to have completely lost its mitocho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine Protease
Serine proteases (or serine endopeptidases) are enzymes that cleave peptide bonds in proteins. Serine serves as the nucleophilic amino acid at the (enzyme's) active site. They are found ubiquitously in both eukaryotes and prokaryotes. Serine proteases fall into two broad categories based on their structure: chymotrypsin-like (trypsin-like) or subtilisin-like. Classification The MEROPS protease classification system counts 16 superfamilies (as of 2013) each containing many families. Each superfamily uses the catalytic triad or dyad in a different protein fold and so represent convergent evolution of the catalytic mechanism. The majority belong to the S1 family of the PA clan (superfamily) of proteases. For superfamilies, P: superfamily, containing a mixture of nucleophile class families, S: purely serine proteases. superfamily. Within each superfamily, families are designated by their catalytic nucleophile, (S: serine proteases). Substrate specificity Serine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-Jun N-terminal Kinase
c-Jun N-terminal kinases (JNKs), were originally identified as kinases that bind and phosphorylate c-Jun on Ser-63 and Ser-73 within its transcriptional activation domain. They belong to the mitogen-activated protein kinase family, and are responsive to stress stimuli, such as cytokines, ultraviolet irradiation, heat shock, and osmotic shock. They also play a role in T cell differentiation and the cellular apoptosis pathway. Activation occurs through a dual phosphorylation of threonine (Thr) and tyrosine (Tyr) residues within a Thr- Pro-Tyr motif located in kinase subdomain VIII. Activation is carried out by two MAP kinase kinases, MKK4 and MKK7, and JNK can be inactivated by Ser/Thr and Tyr protein phosphatases. It has been suggested that this signaling pathway contributes to inflammatory responses in mammals and insects. Isoforms The c-Jun N-terminal kinases consist of ten isoforms derived from three genes: JNK1 (four isoforms), JNK2 (four isoforms) and JNK3 (tw ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
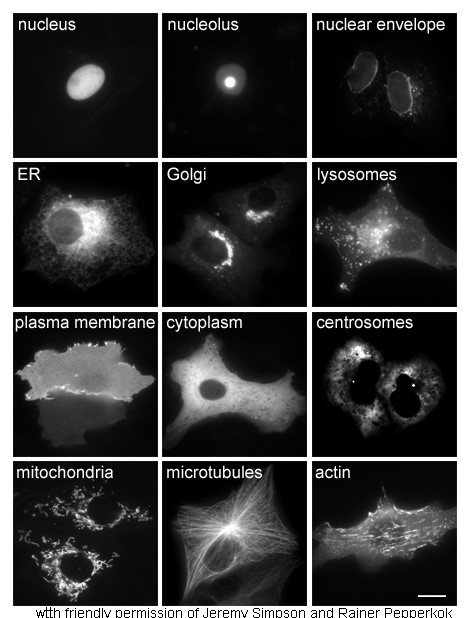
Subcellular Localization
The cells of eukaryotic organisms are elaborately subdivided into functionally-distinct membrane-bound compartments. Some major constituents of eukaryotic cells are: extracellular space, plasma membrane, cytoplasm, nucleus, mitochondria, Golgi apparatus, endoplasmic reticulum (ER), peroxisome, vacuoles, cytoskeleton, nucleoplasm, nucleolus, nuclear matrix and ribosomes. Bacteria also have subcellular localizations that can be separated when the cell is fractionated. The most common localizations referred to include the cytoplasm, the cytoplasmic membrane (also referred to as the inner membrane in Gram-negative bacteria), the cell wall (which is usually thicker in Gram-positive bacteria) and the extracellular environment. The cytoplasm, the cytoplasmic membrane and the cell wall are subcellular localizations, whereas the extracellular environment is clearly not. Most Gram-negative bacteria also contain an outer membrane and periplasmic space. Unlike eukaryotes, most bacteria cont ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. The main components of the cytoplasm are cytosol (a gel-like substance), the organelles (the cell's internal sub-structures), and various cytoplasmic inclusions. The cytoplasm is about 80% water and is usually colorless. The submicroscopic ground cell substance or cytoplasmic matrix which remains after exclusion of the cell organelles and particles is groundplasm. It is the hyaloplasm of light microscopy, a highly complex, polyphasic system in which all resolvable cytoplasmic elements are suspended, including the larger organelles such as the ribosomes, mitochondria, the plant plastids, lipid droplets, and vacuoles. Most cellular activities take place within the cytoplasm, such as many metabolic pathways including glycolysis, and proces ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell Nucleus
The cell nucleus (pl. nuclei; from Latin or , meaning ''kernel'' or ''seed'') is a membrane-bound organelle found in eukaryotic cells. Eukaryotic cells usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have no nuclei, and a few others including osteoclasts have many. The main structures making up the nucleus are the nuclear envelope, a double membrane that encloses the entire organelle and isolates its contents from the cellular cytoplasm; and the nuclear matrix, a network within the nucleus that adds mechanical support. The cell nucleus contains nearly all of the cell's genome. Nuclear DNA is often organized into multiple chromosomes – long stands of DNA dotted with various proteins, such as histones, that protect and organize the DNA. The genes within these chromosomes are structured in such a way to promote cell function. The nucleus maintains the integrity of genes and controls the activities of the cell by regulating gene expres ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Archaebacteria
Archaea ( ; singular archaeon ) is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaebacteria kingdom), but this term has fallen out of use. Archaeal cells have unique properties separating them from the other two domains, Bacteria and Eukaryota. Archaea are further divided into multiple recognized phyla. Classification is difficult because most have not been isolated in a laboratory and have been detected only by their gene sequences in environmental samples. Archaea and bacteria are generally similar in size and shape, although a few archaea have very different shapes, such as the flat, square cells of ''Haloquadratum walsbyi''. Despite this morphological similarity to bacteria, archaea possess genes and several metabolic pathways that are more closely related to those of eukaryotes, notably for the enzymes involved in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacteria and Archaea (both prokaryotes) make up the other two domains. The eukaryotes are usually now regarded as having emerged in the Archaea or as a sister of the Asgard archaea. This implies that there are only two domains of life, Bacteria and Archaea, with eukaryotes incorporated among archaea. Eukaryotes represent a small minority of the number of organisms, but, due to their generally much larger size, their collective global biomass is estimated to be about equal to that of prokaryotes. Eukaryotes emerged approximately 2.3–1.8 billion years ago, during the Proterozoic eon, likely as flagellated phagotrophs. Their name comes from the Greek εὖ (''eu'', "well" or "good") and κάρυον (''karyon'', "nut" or "kernel"). Euka ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |






.jpg)