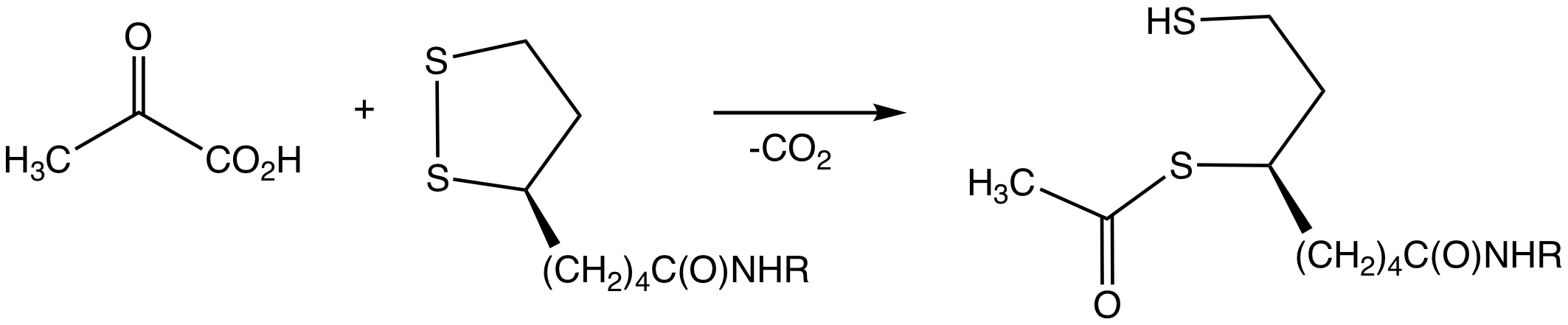
|
Compartment (chemistry)
In chemistry, a compartment is a part of a protein that serves a specific function. They are essentially protein subunits with the added condition that a compartment has distinct functionality, rather than being just a structural component. There may be multiple compartments on one and the same protein. One example is the case of Pyruvate dehydrogenase complex. This is the enzyme which catalyses Pyruvate decarboxylation, the reaction of Pyruvate with Coenzyme A and the major entry point into the TCA cycle: :Pyruvate + Coenzyme A + NAD+ ⇒ acetyl-CoA + NADH + H+ + CO2 Pyruvate dehydrogenase has three chemical compartments; E1 ( pyruvate decarboxylase), E2 (dihydrolipoyl transacetylase Dihydrolipoyl transacetylase (or dihydrolipoamide acetyltransferase) is an enzyme component of the multienzyme pyruvate dehydrogenase complex. The pyruvate dehydrogenase complex is responsible for the pyruvate decarboxylation step that links glyco ...) and E3 ( dihydrolipoyl dehydrogenase). Each one o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemistry
Chemistry is the science, scientific study of the properties and behavior of matter. It is a natural science that covers the Chemical element, elements that make up matter to the chemical compound, compounds made of atoms, molecules and ions: their composition, structure, properties, behavior and the changes they undergo during a Chemical reaction, reaction with other Chemical substance, substances. Chemistry also addresses the nature of chemical bonds in chemical compounds. In the scope of its subject, chemistry occupies an intermediate position between physics and biology. It is sometimes called the central science because it provides a foundation for understanding both Basic research, basic and Applied science, applied scientific disciplines at a fundamental level. For example, chemistry explains aspects of plant growth (botany), the formation of igneous rocks (geology), how atmospheric ozone is formed and how environmental pollutants are degraded (ecology), the properties ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Subunit
In structural biology, a protein subunit is a polypeptide chain or single protein molecule that assembles (or "''coassembles''") with others to form a protein complex. Large assemblies of proteins such as viruses often use a small number of types of protein subunits as building blocks. A subunit is often named with a Greek or Roman letter, and the numbers of this type of subunit in a protein is indicated by a subscript. For example, ATP synthase has a type of subunit called α. Three of these are present in the ATP synthase molecule, leading to the designation α3. Larger groups of subunits can also be specified, like α3β3-hexamer and c-ring. Naturally-occurring proteins that have a relatively small number of subunits are referred to as oligomeric.Quote: ''Oligomer molecule: A molecule of intermediate relative molecular mass, the structure of which essentially comprises a small plurality of units derived, actually or conceptually, from molecules of lower relative molecular ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyruvate Dehydrogenase Complex
Pyruvate dehydrogenase complex (PDC) is a complex of three enzymes that converts pyruvate into acetyl-CoA by a process called pyruvate decarboxylation. Acetyl-CoA may then be used in the citric acid cycle to carry out cellular respiration, and this complex links the glycolysis metabolic pathway to the citric acid cycle. Pyruvate decarboxylation is also known as the "pyruvate dehydrogenase reaction" because it also involves the oxidation of pyruvate. This multi-enzyme complex is related structurally and functionally to the oxoglutarate dehydrogenase and branched-chain oxo-acid dehydrogenase multi-enzyme complexes. Reaction The reaction catalysed by pyruvate dehydrogenase complex is: Structure Pyruvate dehydrogenase (E1) The E1 subunit, called the pyruvate dehydrogenase subunit, has a structure that consists of two chains (an “ɑ” and “ꞵ” chain). A magnesium ion forms a 4-coordinate complex with three, polar amino acid residues (Asp, Asn, and Tyr) located on t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyruvate Decarboxylation
Pyruvate decarboxylation or pyruvate oxidation, also known as the link reaction (or oxidative decarboxylation of pyruvate), is the conversion of pyruvate into acetyl-CoA by the enzyme complex pyruvate dehydrogenase complex. The reaction may be simplified as: :Pyruvate + NAD+ + CoA → Acetyl-CoA + NADH + CO2 Pyruvate oxidation is the step that connects glycolysis and the Krebs cycle. In glycolysis, a single glucose molecule (6 carbons) is split into 2 pyruvates (3 carbons each). Because of this, the link reaction occurs twice for each glucose molecule to produce a total of 2 acetyl-CoA molecules, which can then enter the Krebs cycle. Energy-generating ions and molecules, such as amino acids and carbohydrates, enter the Krebs cycle as acetyl coenzyme A and oxidize in the cycle. The pyruvate dehydrogenase complex (PDC) catalyzes the decarboxylation of pyruvate, resulting in the synthesis of acetyl-CoA, CO2, and NADH. In eukaryotes, this enzyme complex regulates pyruvate metabo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TCA Cycle
The citric acid cycle (CAC)—also known as the Krebs cycle or the TCA cycle (tricarboxylic acid cycle)—is a series of chemical reactions to release stored energy through the oxidation of acetyl-CoA derived from carbohydrates, fats, and proteins. The Krebs cycle is used by organisms that respire (as opposed to organisms that ferment) to generate energy, either by anaerobic respiration or aerobic respiration. In addition, the cycle provides precursors of certain amino acids, as well as the reducing agent NADH, that are used in numerous other reactions. Its central importance to many biochemical pathways suggests that it was one of the earliest components of metabolism and may have originated abiogenically. Even though it is branded as a 'cycle', it is not necessary for metabolites to follow only one specific route; at least three alternative segments of the citric acid cycle have been recognized. The name of this metabolic pathway is derived from the citric acid (a tricarboxy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyruvate Dehydrogenase
Pyruvate dehydrogenase is an enzyme that catalyzes the reaction of pyruvate and a lipoamide to give the acetylated dihydrolipoamide and carbon dioxide. The conversion requires the coenzyme thiamine pyrophosphate. Pyruvate dehydrogenase is usually encountered as a component, referred to as E1, of the pyruvate dehydrogenase complex (PDC). PDC consists of other enzymes, referred to as E2 and E3. Collectively E1-E3 transform pyruvate, NAD+, coenzyme A into acetyl-CoA, CO2, and NADH. The conversion is crucial because acetyl-CoA may then be used in the citric acid cycle to carry out cellular respiration. To distinguish between this enzyme and the PDC, it is systematically called pyruvate dehydrogenase (acetyl-transferring). Mechanism The thiamine pyrophosphate (TPP) converts to an ylide by deprotonation. The ylide attack the ketone group of pyruvate. The resulting adduct decarboxylates. The resulting 1,3-dipole reductively acetylates lipoamide-E2. In terms of details, bioch ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyruvate Decarboxylase
Pyruvate decarboxylase is an enzyme () that catalyses the decarboxylation of pyruvic acid to acetaldehyde. It is also called 2-oxo-acid carboxylase, alpha-ketoacid carboxylase, and pyruvic decarboxylase. In anaerobic conditions, this enzyme is participates in the fermentation process that occurs in yeast, especially of the genus ''Saccharomyces'', to produce ethanol by fermentation. It is also present in some species of fish (including goldfish and carp) where it permits the fish to perform ethanol fermentation (along with lactic acid fermentation) when oxygen is scarce. Pyruvate decarboxylase starts this process by converting pyruvate into acetaldehyde and carbon dioxide. Pyruvate decarboxylase depends on cofactors thiamine pyrophosphate (TPP) and magnesium. This enzyme should not be mistaken for the unrelated enzyme pyruvate dehydrogenase, an oxidoreductase (), that catalyzes the oxidative decarboxylation of pyruvate to acetyl-CoA. Structure Pyruvate decarboxylase occurs a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dihydrolipoyl Transacetylase
Dihydrolipoyl transacetylase (or dihydrolipoamide acetyltransferase) is an enzyme component of the multienzyme pyruvate dehydrogenase complex. The pyruvate dehydrogenase complex is responsible for the pyruvate decarboxylation step that links glycolysis to the citric acid cycle. This involves the transformation of pyruvate from glycolysis into acetyl-CoA which is then used in the citric acid cycle to carry out cellular respiration. There are three different enzyme components in the pyruvate dehydrogenase complex. Pyruvate dehydrogenase (EC 1.2.4.1) is responsible for the oxidation of pyruvate, dihydrolipoyl transacetylase (this enzyme; EC 2.3.1.12) transfers the acetyl group to coenzyme A (CoA), and dihydrolipoyl dehydrogenase (EC 1.8.1.4) regenerates the lipoamide. Because dihydrolipoyl transacetylase is the second of the three enzyme components participating in the reaction mechanism for conversion of pyruvate into acetyl CoA, it is sometimes referred to as E2. In humans, dihydrol ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dihydrolipoyl Dehydrogenase
Dihydrolipoamide dehydrogenase (DLD), also known as dihydrolipoyl dehydrogenase, mitochondrial, is an enzyme that in humans is encoded by the ''DLD'' gene. DLD is a flavoprotein enzyme that oxidizes dihydrolipoamide to lipoamide. Dihydrolipoamide dehydrogenase (DLD) is a mitochondrial enzyme that plays a vital role in energy metabolism in eukaryotes. This enzyme is required for the complete reaction of at least five different multi-enzyme complexes. Additionally, DLD is a flavoenzyme oxidoreductase that contains a reactive disulfide bridge and a FAD cofactor that are directly involved in catalysis. The enzyme associates into tightly bound homodimers required for its enzymatic activity. File:Lipoamide-2D-skeletal.png, Lipoamide File:Dihydrolipoamide.svg, Dihydrolipoamide Structure The protein encoded by the DLD gene comes together with another protein to form a dimer in the central metabolic pathway. Several amino acids within the catalytic pocket have been identified as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |