|
CHD2
Chromodomain-helicase-DNA-binding protein 2 is an enzyme that in humans is encoded by the ''CHD2'' gene. Function The CHD family of proteins is characterized by the presence of chromo (chromatin organization modifier) domains and SNF2-related helicase/ATPase domains. CHD genes alter gene expression possibly by modification of chromatin structure thus altering access of the transcriptional apparatus to its chromosomal DNA template. CHD2 catalyzes the assembly of chromatin into periodic arrays; and the N-terminal region of CHD2, which contains tandem chromodomains, serves an auto-inhibitory role in both the DNA-binding and ATPase activities of CHD2. Alternatively spliced transcript variants encoding distinct isoforms have been found for this gene. Clinical significance De novo mutations and deletions in this gene have been associated with cases of epileptic encephalopathies. CHD2 epilepsy is increasingly being identified as a subpopulation of Lennox-Gastaut Syndrome. CHD ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromodomain Helicase DNA-binding (CHD) Protein Subfamily
Chromodomain helicase DNA-binding (CHD) proteins is a subfamily of ATP-dependent chromatin remodeling complexes (remodelers). All remodelers fall under the umbrella of RNA/DNA helicase superfamily 2. In yeast, CHD complexes are primarily responsible for nucleosome assembly and organization. These complexes play an additional role in multicellular eukaryotes, assisting in chromatin access and nucleosome editing. Functions of CHD subfamily proteins Similar to the imitation switch (ISWI) subfamily of ATP-dependent chromatin remodelers, CHD complexes regulate the assembly and organization of mature nucleosomes along the DNA. Histones are removed during DNA replication; following behind the replisome, histones start to assemble as immature pre-nucleosomes on nascent DNA. With the help of CHD complexes, histone octamers can mature into native nucleosomes. Following nucleosome formation, CHD complexes organize nucleosomes by regularly spacing them apart along the DNA. Additionally, CHDs ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromodomain
A chromodomain (''chromatin organization modifier'') is a protein structural domain of about 40–50 amino acid residues commonly found in proteins associated with the remodeling and manipulation of chromatin. The domain is highly conserved among both plants and animals, and is represented in a large number of different proteins in many genomes, such as that of the mouse. Some chromodomain-containing genes have multiple alternative splicing isoforms that omit the chromodomain entirely. In mammals, chromodomain-containing proteins are responsible for aspects of gene regulation related to chromatin remodeling and formation of heterochromatin regions. Chromodomain-containing proteins also bind methylated histones and appear in the RNA-induced transcriptional silencing RNA-induced transcriptional silencing (RITS) is a form of RNA interference by which short RNA molecules – such as small interfering RNA (siRNA) – trigger the downregulation of transcription of a particular g ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SMARCA2
Probable global transcription activator SNF2L2 is a protein that in humans is encoded by the ''SMARCA2'' gene. Function The protein encoded by this gene is a member of the SWI/SNF family of proteins and is highly similar to the brahma protein of Drosophila. Members of this family have helicase and ATPase activities and are thought to regulate transcription of certain genes by altering the chromatin structure around those genes. The encoded protein is part of the large ATP-dependent chromatin remodeling complex SNF/SWI, which is required for transcriptional activation of genes normally repressed by chromatin. Two transcript variants encoding different isoforms have been found for this gene, which contains a trinucleotide repeat (CAG) length polymorphism. Interactions SMARCA2 has been shown to interact with: * ACTL6A, * ARID1B, * CEBPB, * POLR2A, * Prohibitin, * SIN3A, * SMARCB1, * SMARCC1, and * SS18 Protein SSXT is a protein that in humans is encoded by the ''SS18'' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ATPase
ATPases (, Adenosine 5'-TriPhosphatase, adenylpyrophosphatase, ATP monophosphatase, triphosphatase, SV40 T-antigen, ATP hydrolase, complex V (mitochondrial electron transport), (Ca2+ + Mg2+)-ATPase, HCO3−-ATPase, adenosine triphosphatase) are a class of enzymes that catalyze the decomposition of ATP into ADP and a free phosphate ion or the inverse reaction. This dephosphorylation reaction releases energy, which the enzyme (in most cases) harnesses to drive other chemical reactions that would not otherwise occur. This process is widely used in all known forms of life. Some such enzymes are integral membrane proteins (anchored within biological membranes), and move solutes across the membrane, typically against their concentration gradient. These are called transmembrane ATPases. Functions Transmembrane ATPases import metabolites necessary for cell metabolism and export toxins, wastes, and solutes that can hinder cellular processes. An important example is the sodium-potass ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromatin
Chromatin is a complex of DNA and protein found in eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in reinforcing the DNA during cell division, preventing DNA damage, and regulating gene expression and DNA replication. During mitosis and meiosis, chromatin facilitates proper segregation of the chromosomes in anaphase; the characteristic shapes of chromosomes visible during this stage are the result of DNA being coiled into highly condensed chromatin. The primary protein components of chromatin are histones. An octamer of two sets of four histone cores (Histone H2A, Histone H2B, Histone H3, and Histone H4) bind to DNA and function as "anchors" around which the strands are wound.Maeshima, K., Ide, S., & Babokhov, M. (2019). Dynamic chromatin organization without the 30-nm fiber. ''Current opinion in cell biology, 58,'' 95–104. https://doi.o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
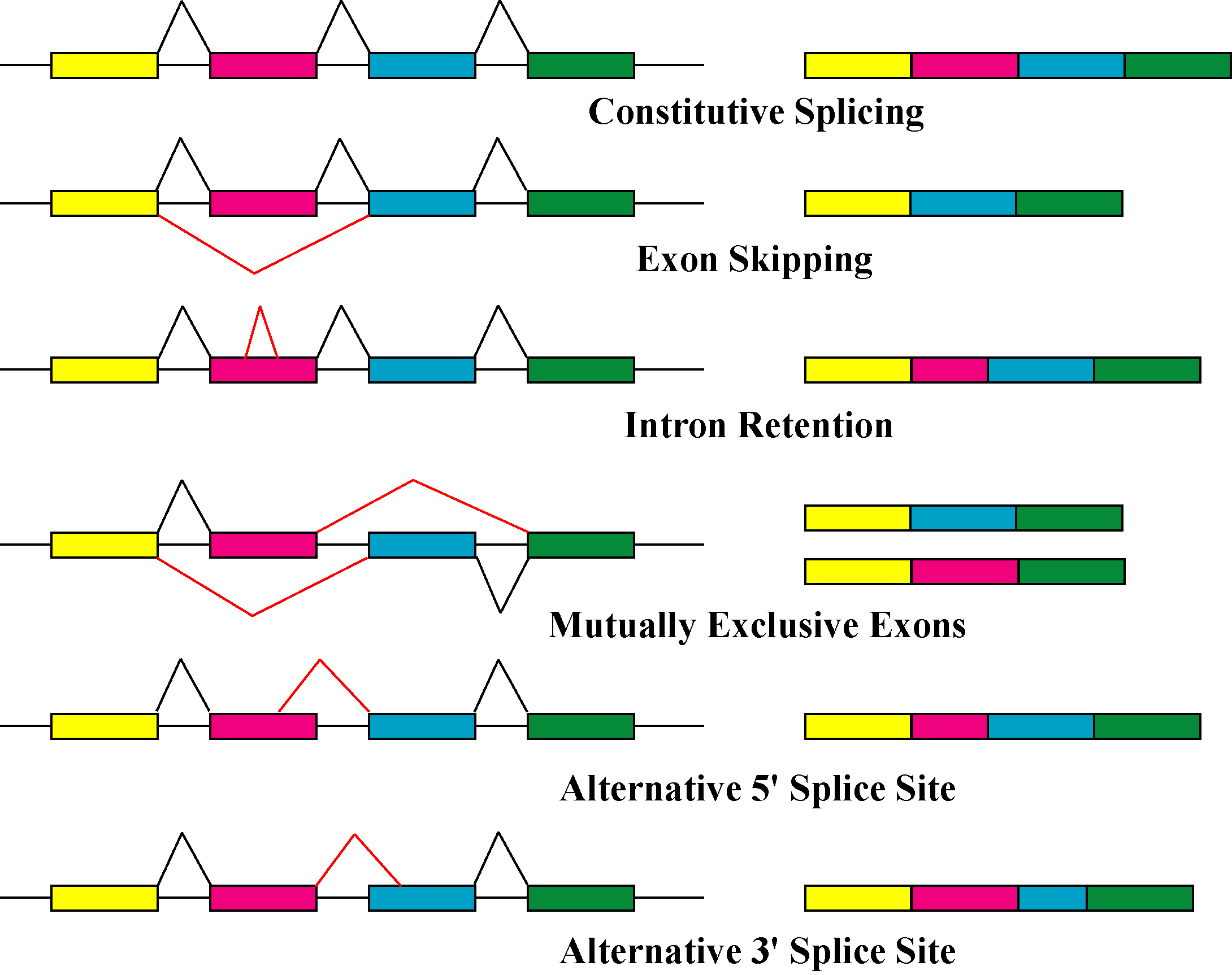
Alternative Splicing
Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. This means the exons are joined in different combinations, leading to different (alternative) mRNA strands. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions (see Figure). Biologically relevant alternative splicing occurs as a normal phenomenon in eukaryotes, where it increases the number of proteins that can be encoded by the genome. In humans, it is widely believed that ~95% of multi-exonic genes are alternatively spliced to produce functional alternative products from the same gene but many scientists believe that most o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Isoform
A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. (For that, see Proteoforms.) Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments ( exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein. The discovery of isoforms could explain the discrepancy between the small number of protein coding regions genes revealed by the human genome project and the large diversity of proteins seen in an organism: different ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CHD8
Chromodomain-helicase-DNA-binding protein 8 is an enzyme that in humans is encoded by the ''CHD8'' gene. Function The gene CHD8 encodes the protein chromodomain helicase DNA binding protein 8, which is a chromatin regulator enzyme that is essential during fetal development. CHD8 is an ATP dependent enzyme. The protein contains an Snf2 helicase domain that is responsible for the hydrolysis of ATP to ADP. CHD8 encodes for a DNA helicase that function as a transcription repressor by remodeling chromatin structure by altering the position of nucleosomes. CHD8 negatively regulates Wnt signaling. Wnt signaling is important in the vertebrate early development and morphogenesis. It is believed that CHD8 also recruits the linker histone H1 and causes the repression of β-catenin and p53 target genes. The importance of CHD8 can be observed in studies where CHD8-knockout mice died after 5.5 embryonic days because of widespread p53 induced apoptosis. Recently CD8 has been associated to th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DYRK1A
Dual specificity tyrosine-phosphorylation-regulated kinase 1A is an enzyme that in humans is encoded by the ''DYRK1A'' gene. Alternative splicing of this gene generates several transcript variants differing from each other either in the 5' UTR or in the 3' coding region. These variants encode at least five different isoforms. Function DYRK1A is a member of the dual-specificity tyrosine phosphorylation-regulated kinase (DYRK) family. This member contains a nuclear targeting signal sequence, a protein kinase domain, a leucine zipper motif, and a highly conservative 13-consecutive-histidine repeat. It catalyzes its autophosphorylation on serine/threonine and tyrosine residues. It may play a significant role in a signaling pathway regulating cell proliferation and may be involved in brain development. This gene is a homolog of ''Drosophila'' mnb (minibrain) gene. Dyrk1a has also been shown to modulate plasma homocysteine level in a mouse model of overexpression. Clinical signi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |