|
C3orf14-Chromosome 3 Open Reading Frame 14
The human gene Chromosome 3 open reading frame 14 is a gene of uncertain function located at 3p14.2 near fragile site FRBA3—which falls between this gene and the centromere. Its protein is expected to localize to the nucleus and bind DNA. Orthologs have been identified in all of the major animal groups, minus amphibians and insects, tracing as far back as the sea anemone; indicating an origin of over 1000 mya, highlighting its importance in the animal genome. Gene aliases C3orf14 is also known by the aliases LOC57415, FLJ94553 and FLJ17473. Gene orthologs found in other organisms are usually known by the name c3orf14-like, though some are known as LOC57415-like or HT021-like (protein name). Structure The mRNA is composed of 6 exons, and encodes a 15007.84 kD protein known as HT021. This protein has a pre-modification isoelectric point of 5.57 and alpha helices span most of its length. Four sites of possible phosphorylation have been identified, and at least two sites of ph ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
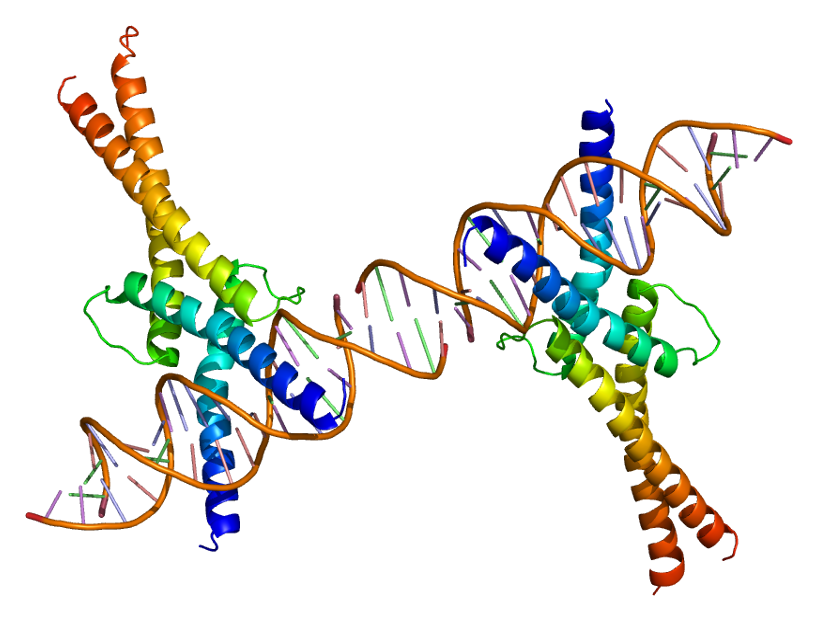
Leucine Zipper
A leucine zipper (or leucine scissors) is a common three-dimensional structural motif in proteins. They were first described by Landschulz and collaborators in 1988 when they found that an enhancer binding protein had a very characteristic 30-amino acid segment and the display of these amino acid sequences on an idealized alpha helix revealed a periodic repetition of leucine residues at every seventh position over a distance covering eight helical turns. The polypeptide segments containing these periodic arrays of leucine residues were proposed to exist in an alpha-helical conformation and the leucine side chains from one alpha helix interdigitate with those from the alpha helix of a second polypeptide, facilitating dimerization. Leucine zippers are a dimerization motif of the bZIP (Basic-region leucine zipper) class of eukaryotic transcription factors. The bZIP domain is 60 to 80 amino acids in length with a highly conserved DNA binding basic region and a more diversified leucin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fish
Fish are aquatic, craniate, gill-bearing animals that lack limbs with digits. Included in this definition are the living hagfish, lampreys, and cartilaginous and bony fish as well as various extinct related groups. Approximately 95% of living fish species are ray-finned fish, belonging to the class Actinopterygii, with around 99% of those being teleosts. The earliest organisms that can be classified as fish were soft-bodied chordates that first appeared during the Cambrian period. Although they lacked a true spine, they possessed notochords which allowed them to be more agile than their invertebrate counterparts. Fish would continue to evolve through the Paleozoic era, diversifying into a wide variety of forms. Many fish of the Paleozoic developed external armor that protected them from predators. The first fish with jaws appeared in the Silurian period, after which many (such as sharks) became formidable marine predators rather than just the prey of arthropods. Mos ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Housekeeping Gene
In molecular biology, housekeeping genes are typically constitutive genes that are required for the maintenance of basic cellular function, and are expressed in all cells of an organism under normal and patho-physiological conditions. Although some housekeeping genes are expressed at relatively constant rates in most non-pathological situations, the expression of other housekeeping genes may vary depending on experimental conditions. The origin of the term "housekeeping gene" remains obscure. Literature from 1976 used the term to describe specifically tRNA and rRNA. For experimental purposes, the expression of one or multiple housekeeping genes is used as a reference point for the analysis of expression levels of other genes. The key criterion for the use of a housekeeping gene in this manner is that the chosen housekeeping gene is uniformly expressed with low variance under both control and experimental conditions. Validation of housekeeping genes should be performed before th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromosomal Fragile Site
A chromosomal fragile site is a specific heritable point on a chromosome that tends to form a gap or constriction and may tend to break when the cell is exposed to partial replication stress. Based on their frequency, fragile sites are classified as "common" or "rare". To date, more than 120 fragile sites have been identified in the human genome. Common fragile sites are considered part of normal chromosome structure and are present in all (or nearly all) individuals in a population. Under normal conditions, most common fragile sites are not prone to spontaneous breaks. Common fragile sites are of interest in cancer studies because they are frequently affected in cancer and they can be found in healthy individuals. Sites FRA3B (harboring the ''FHIT'' gene) and FRA16D (harboring the ''WWOX'' gene) are two well known examples and have been a major focus of research. Rare fragile sites are found in less than 5% of the population, and are often composed of two- or three-nucleotide rep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FHIT
Bis(5'-adenosyl)-triphosphatase also known as fragile histidine triad protein (FHIT) is an enzyme that in humans is encoded by the ''FHIT'' gene. Function FHIT is also known as human accelerated region 10. It may, therefore, have played a key role in differentiating humans from apes. This gene, a member of the histidine triad gene family, encodes a diadenosine P1,P3-bis(5'-adenosyl)-triphosphate adenylohydrolase involved in purine metabolism. The gene encompasses the common fragile site FRA3B on chromosome 3, where carcinogen-induced damage can lead to translocations and aberrant transcripts of this gene. In fact, aberrant transcripts from this gene have been found in about half of all esophageal, stomach, and colon carcinomas. Though the exact molecular function of FHIT is still partially unclear, the gene works as a tumor suppressor as it has been demonstrated in animal studies. Furthermore FHIT has been shown to synergize with VHL, another tumor suppressor, in protectin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tumor Suppressor Gene
A tumor suppressor gene (TSG), or anti-oncogene, is a gene that regulates a cell during cell division and replication. If the cell grows uncontrollably, it will result in cancer. When a tumor suppressor gene is mutated, it results in a loss or reduction in its function. In combination with other genetic mutations, this could allow the cell to grow abnormally. The loss of function for these genes may be even more significant in the development of human cancers, compared to the activation of oncogenes. TSGs can be grouped into the following categories: caretaker genes, gatekeeper genes, and more recently landscaper genes. Caretaker genes ensure stability of the genome via DNA repair and subsequently when mutated allow mutations to accumulate. Meanwhile, gatekeeper genes directly regulate cell growth by either inhibiting cell cycle progression or inducing apoptosis. Lastly landscaper genes regulate growth by contributing to the surrounding environment, when mutated can cause an envir ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetic Tree
A phylogenetic tree (also phylogeny or evolutionary tree Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA.) is a branching diagram or a tree showing the evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical or genetic characteristics. All life on Earth is part of a single phylogenetic tree, indicating common ancestry. In a ''rooted'' phylogenetic tree, each node with descendants represents the inferred most recent common ancestor of those descendants, and the edge lengths in some trees may be interpreted as time estimates. Each node is called a taxonomic unit. Internal nodes are generally called hypothetical taxonomic units, as they cannot be directly observed. Trees are useful in fields of biology such as bioinformatics, systematics, and phylogenetics. ''Unrooted'' trees illustrate only the relatedness of the leaf nodes and do not require the ancestral root to b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Orthologs
Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal (or lateral) gene transfer event (xenologs). Homology among DNA, RNA, or proteins is typically inferred from their nucleotide or amino acid sequence similarity. Significant similarity is strong evidence that two sequences are related by evolutionary changes from a common ancestral sequence. Alignments of multiple sequences are used to indicate which regions of each sequence are homologous. Identity, similarity, and conservation The term "percent homology" is often used to mean "sequence similarity”, that is the percentage of identical residues (''percent identity''), or the percentage of residues conserved with similar physicochemical properties ('' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Anolis Carolinensis
''Anolis carolinensis'' or green anole () (among other names below) is a tree-dwelling species of anole lizard native to the southeastern United States and introduced to islands in the Pacific and Caribbean. A small to medium-sized lizard, the green anole is a trunk-crown ecomorph and can change its color to several shades from brown to green. Other common names include the Carolina anole, Carolina green anole, American anole, American green anole, North American green anole and red-throated anole. It is sometimes referred to as the American chameleon (typically in the pet trade) due to its color-changing ability; however, it is not a true chameleon. Description The green anole is a small to medium-sized lizard, with a slender body. The head is long and pointed with ridges between the eyes and nostrils, and smaller ones on the top of the head. The toes have adhesive pads to facilitate climbing. They exhibit sexual dimorphism, the males being fifteen percent larger. Adult ma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Model Organisms
A model organism (often shortened to model) is a non-human species that is extensively studied to understand particular biological phenomena, with the expectation that discoveries made in the model organism will provide insight into the workings of other organisms. Model organisms are widely used to research human disease when human experimentation would be unfeasible or unethical. This strategy is made possible by the common descent of all living organisms, and the conservation of metabolic and developmental pathways and genetic material over the course of evolution. Studying model organisms can be informative, but care must be taken when generalizing from one organism to another. In researching human disease, model organisms allow for better understanding the disease process without the added risk of harming an actual human. The species chosen will usually meet a determined taxonomic equivalency to humans, so as to react to disease or its treatment in a way that resembles hu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |



_by_Robert_Michniewicz.jpg)
