|
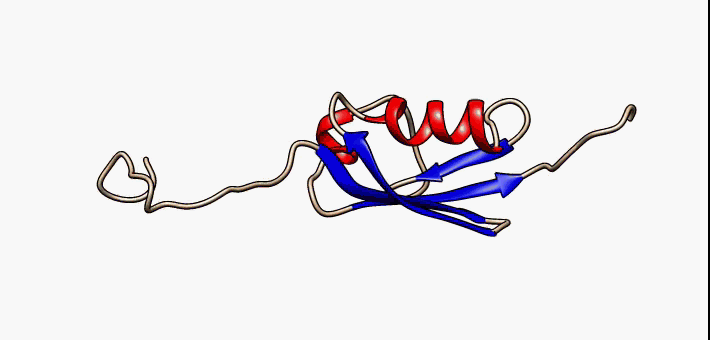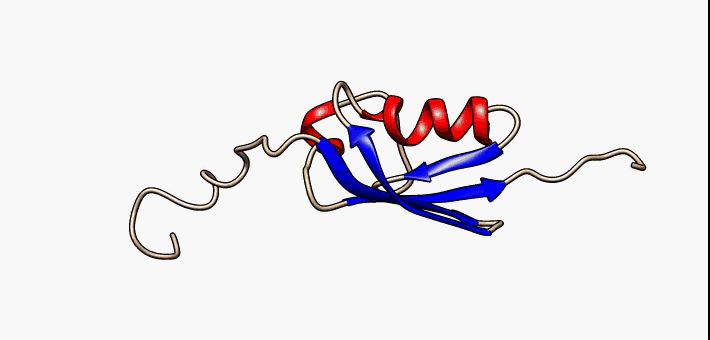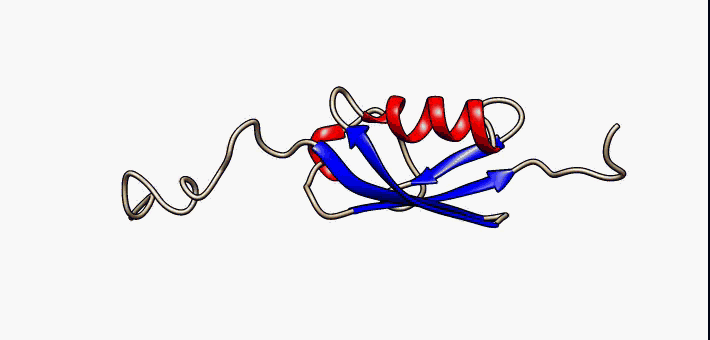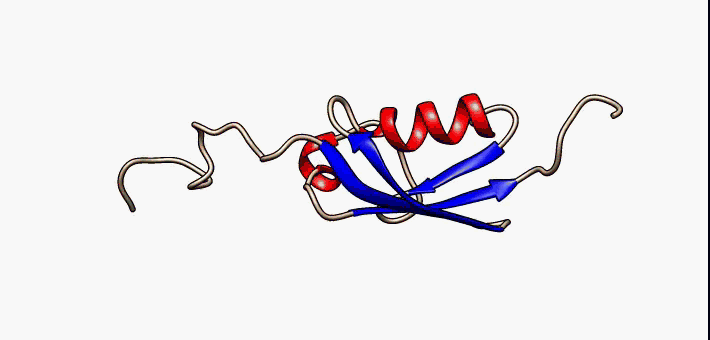
C1orf131 Overview1
Uncharacterized protein C1orf131 is a protein that in humans is encoded by the gene ''C1orf131''. The first ortholog of this protein was discovered in humans. Subsequently, through the use of algorithms and bioinformatics, homologs of C1orf131 have been discovered in numerous species, and as a result, the name of the majority of the proteins in this protein family is Uncharacterized protein C1orf131 homolog. Gene In humans ''C1orf131'' is located on the minus strand of chromosome 1 and on the cytogenetic band 1q42.2 along with 193 other genes. Notably, the gene upstream of ''C1orf131'' is ''GNPAT'', and the gene downstream of ''C1orf131'' is Tripartite motif family, ''TRIM67''. When this gene is transcribed in humans, ''C1orf131'' most often forms an mRNA of 1458 base pairs long which is composed of seven exons. There are at least nine others alternative splice forms in humans that produce proteins. They range in size from 129 base pairs (2 exons) to 1458 base pairs (7 exons). ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A. The addition of ubiquitin to a substrate protein is called ubiquitylation (or, alternatively, ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide bond, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Evolutionary History Of C1orf131
Evolution is change in the heritable characteristics of biological populations over successive generations. These characteristics are the expressions of genes, which are passed on from parent to offspring during reproduction. Variation tends to exist within any given population as a result of genetic mutation and recombination. Evolution occurs when evolutionary processes such as natural selection (including sexual selection) and genetic drift act on this variation, resulting in certain characteristics becoming more common or more rare within a population. The evolutionary pressures that determine whether a characteristic is common or rare within a population constantly change, resulting in a change in heritable characteristics arising over successive generations. It is this process of evolution that has given rise to biodiversity at every level of biological organisation, including the levels of species, individual organisms, and molecules. The theory of evolution by na ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fibrinogen
Fibrinogen (factor I) is a glycoprotein complex, produced in the liver, that circulates in the blood of all vertebrates. During tissue and vascular injury, it is converted enzymatically by thrombin to fibrin and then to a fibrin-based blood clot. Fibrin clots function primarily to occlude blood vessels to stop bleeding. Fibrin also binds and reduces the activity of thrombin. This activity, sometimes referred to as antithrombin I, limits clotting. Fibrin also mediates blood platelet and endothelial cell spreading, tissue fibroblast proliferation, capillary tube formation, and angiogenesis and thereby promotes revascularization and wound healing. Reduced and/or dysfunctional fibrinogens occur in various congenital and acquired human fibrinogen-related disorders. These disorders represent a group of rare conditions in which individuals may present with severe episodes of pathological bleeding and thrombosis; these conditions are treated by supplementing blood fibrinogen levels an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pseudogenes
Pseudogenes are nonfunctional segments of DNA that resemble functional genes. Most arise as superfluous copies of functional genes, either directly by DNA duplication or indirectly by reverse transcription of an mRNA transcript. Pseudogenes are usually identified when genome sequence analysis finds gene-like sequences that lack regulatory sequences needed for transcription or translation, or whose coding sequences are obviously defective due to frameshifts or premature stop codons. Most non-bacterial genomes contain many pseudogenes, often as many as functional genes. This is not surprising, since various biological processes are expected to accidentally create pseudogenes, and there are no specialized mechanisms to remove them from genomes. Eventually pseudogenes may be deleted from their genomes by chance DNA replication or DNA repair errors, or they may accumulate so many mutational changes that they are no longer recognizable as former genes. Analysis of these degeneration ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intrinsically Disordered Protein
In molecular biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered three-dimensional structure, typically in the absence of its macromolecular interaction partners, such as other proteins or RNA. IDPs range from fully unstructured to partially structured and include random coil, molten globule-like aggregates, or flexible linkers in large multi-domain proteins. They are sometimes considered as a separate class of proteins along with globular, fibrous and membrane proteins. IDPs are a very large and functionally important class of proteins and their discovery has disproved the idea that three-dimensional structures of proteins must be fixed to accomplish their biological functions. For example, IDPs have been identified to participate in weak multivalent In chemistry, polyvalency (or polyvalence, multivalency) is the property of chemical species (generally atoms or molecules) that exhibit more than one valence by forming multiple chem ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enterococcus Faecalis
''Enterococcus faecalis'' – formerly classified as part of the group D ''Streptococcus'' system – is a Gram-positive, commensal bacterium inhabiting the gastrointestinal tracts of humans. Like other species in the genus ''Enterococcus'', ''E. faecalis'' is found in healthy humans and can be used as a probiotic. The probiotic strains such as Symbioflor1 and EF-2001 are characterized by the lack of specific genes related to drug resistance and pathogenesis. As an opportunistic pathogen, ''E. faecalis'' can cause life-threatening infections, especially in the nosocomial (hospital) environment, where the naturally high levels of antibiotic resistance found in ''E. faecalis'' contribute to its pathogenicity. ''E. faecalis'' has been frequently found in reinfected, root canal-treated teeth in prevalence values ranging from 30% to 90% of the cases. Re-infected root canal-treated teeth are about nine times more likely to harbor ''E. faecalis'' than cases of primary infections. Physi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacillus
''Bacillus'' (Latin "stick") is a genus of Gram-positive, rod-shaped bacteria, a member of the phylum ''Bacillota'', with 266 named species. The term is also used to describe the shape (rod) of other so-shaped bacteria; and the plural ''Bacilli'' is the name of the class of bacteria to which this genus belongs. ''Bacillus'' species can be either obligate aerobes which are dependent on oxygen, or facultative anaerobes which can survive in the absence of oxygen. Cultured ''Bacillus'' species test positive for the enzyme catalase if oxygen has been used or is present. ''Bacillus'' can reduce themselves to oval endospores and can remain in this dormant state for years. The endospore of one species from Morocco is reported to have survived being heated to 420 °C. Endospore formation is usually triggered by a lack of nutrients: the bacterium divides within its cell wall, and one side then engulfs the other. They are not true spores (i.e., not an offspring). Endospore formation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Herpesviridae
''Herpesviridae'' is a large family of DNA viruses that cause infections and certain diseases in animals, including humans. The members of this family are also known as herpesviruses. The family name is derived from the Greek word ''ἕρπειν'' ( 'to creep'), referring to spreading cutaneous lesions, usually involving blisters, seen in flares of herpes simplex 1, herpes simplex 2 and herpes zoster ( shingles). In 1971, the International Committee on the Taxonomy of Viruses (ICTV) established ''Herpesvirus'' as a genus with 23 viruses among four groups. As of 2020, 115 species are recognized, all but one of which are in one of the three subfamilies. Herpesviruses can cause both latent and lytic infections. Nine herpesvirus types are known to primarily infect humans, at least five of which – herpes simplex viruses 1 and 2 (HSV-1 and HSV-2, also known as HHV-1 and HHV-2; both of which can cause orolabial herpes and genital herpes), varicella zoster virus (or HHV-3; the cause ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C1orf131 Overview1
Uncharacterized protein C1orf131 is a protein that in humans is encoded by the gene ''C1orf131''. The first ortholog of this protein was discovered in humans. Subsequently, through the use of algorithms and bioinformatics, homologs of C1orf131 have been discovered in numerous species, and as a result, the name of the majority of the proteins in this protein family is Uncharacterized protein C1orf131 homolog. Gene In humans ''C1orf131'' is located on the minus strand of chromosome 1 and on the cytogenetic band 1q42.2 along with 193 other genes. Notably, the gene upstream of ''C1orf131'' is ''GNPAT'', and the gene downstream of ''C1orf131'' is Tripartite motif family, ''TRIM67''. When this gene is transcribed in humans, ''C1orf131'' most often forms an mRNA of 1458 base pairs long which is composed of seven exons. There are at least nine others alternative splice forms in humans that produce proteins. They range in size from 129 base pairs (2 exons) to 1458 base pairs (7 exons). ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amyloid
Amyloids are aggregates of proteins characterised by a Fibril, fibrillar morphology of 7–13 Nanometer, nm in diameter, a beta sheet (β-sheet) Secondary structure of proteins, secondary structure (known as cross-β) and ability to be Staining, stained by particular dyes, such as Congo red. In the human body, amyloids have been linked to the development of various diseases. Pathogenic amyloids form when previously healthy proteins lose their normal Protein structure, structure and physiology, physiological functions (Protein misfolding, misfolding) and form fibrous deposits in amyloid plaques around cells which can disrupt the healthy function of tissues and organs. Such amyloids have been associated with (but not necessarily as the cause of) more than 50 human diseases, known as amyloidosis, and may play a role in some neurodegenerative diseases. Some of these diseases are mainly sporadic and only a few cases are Genetic disorder, familial. Others are only Genetic disorder, fam ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mass Spectrometry
Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a ''mass spectrum'', a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is used in many different fields and is applied to pure samples as well as complex mixtures. A mass spectrum is a type of plot of the ion signal as a function of the mass-to-charge ratio. These spectra are used to determine the elemental or isotopic signature of a sample, the masses of particles and of molecules, and to elucidate the chemical identity or structure of molecules and other chemical compounds. In a typical MS procedure, a sample, which may be solid, liquid, or gaseous, is ionized, for example by bombarding it with a beam of electrons. This may cause some of the sample's molecules to break up into positively charged fragments or simply become positively charged without fragmenting. These ions (fragments) are then separated accordin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |









.jpg)