|
C18orf63
Chromosome 18 open reading frame 63 is a protein which in humans is encoded by the C18orf63 gene. This protein is not yet well understood by the scientific community. Research has been conducted suggesting that C18orf63 could be a potential biomarker for early stage pancreatic cancer and breast cancer. Gene This gene is located at band 22, sub-band 3, on the long arm of chromosome 18. It is composed of 5065 base pairs spanning from 74,315,875 to 74,359,187 bp on chromosome 18. The gene has a total of 14 exons. C18orf63 is also known by the alias DKFZP78G0119. No isoforms exist for this gene. Expression C18orf63 has high expression in the testis. The gene shows low expression in the kidneys, liver, lung, and pelvis. There is no phenotype associated with this gene. Promoter The promoter region for C18orf63 is 1163 bp long starting at 74,314,813 bp and ending at 74,315,975 bp. The promoter ID is GXP_4417391. The presence of multiple y-box binding transcription factors an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromosome 18 (human)
Chromosome 18 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 18 spans about 80 million base pairs (the building material of DNA) and represents about 2.5 percent of the total DNA in cells. Genes Number of genes The following are some of the gene count estimates of human chromosome 18. Because researchers use different approaches to genome annotation their predictions of the number of genes on each chromosome varies (for technical details, see gene prediction). Among various projects, the collaborative consensus coding sequence project ( CCDS) takes an extremely conservative strategy. So CCDS's gene number prediction represents a lower bound on the total number of human protein-coding genes. Gene list The following is a partial list of genes on human chromosome 18. For complete list, see the link in the infobox on the right. Diseases and disorders The following diseases are some of those related to genes o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isoleucine
Isoleucine (symbol Ile or I) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), an α-carboxylic acid group (which is in the deprotonated −COO form under biological conditions), and a hydrocarbon side chain with a branch (a central carbon atom bound to three other carbon atoms). It is classified as a non-polar, uncharged (at physiological pH), branched-chain, aliphatic amino acid. It is essential in humans, meaning the body cannot synthesize it, and must be ingested in our diet. Isoleucine is synthesized from pyruvate employing leucine biosynthesis enzymes in other organisms such as bacteria. It is encoded by the codons AUU, AUC, and AUA. Metabolism Biosynthesis As an essential nutrient, it is not synthesized in the body, hence it must be ingested, usually as a component of proteins. In plants and microorganisms, it is synthesized via several steps, startin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
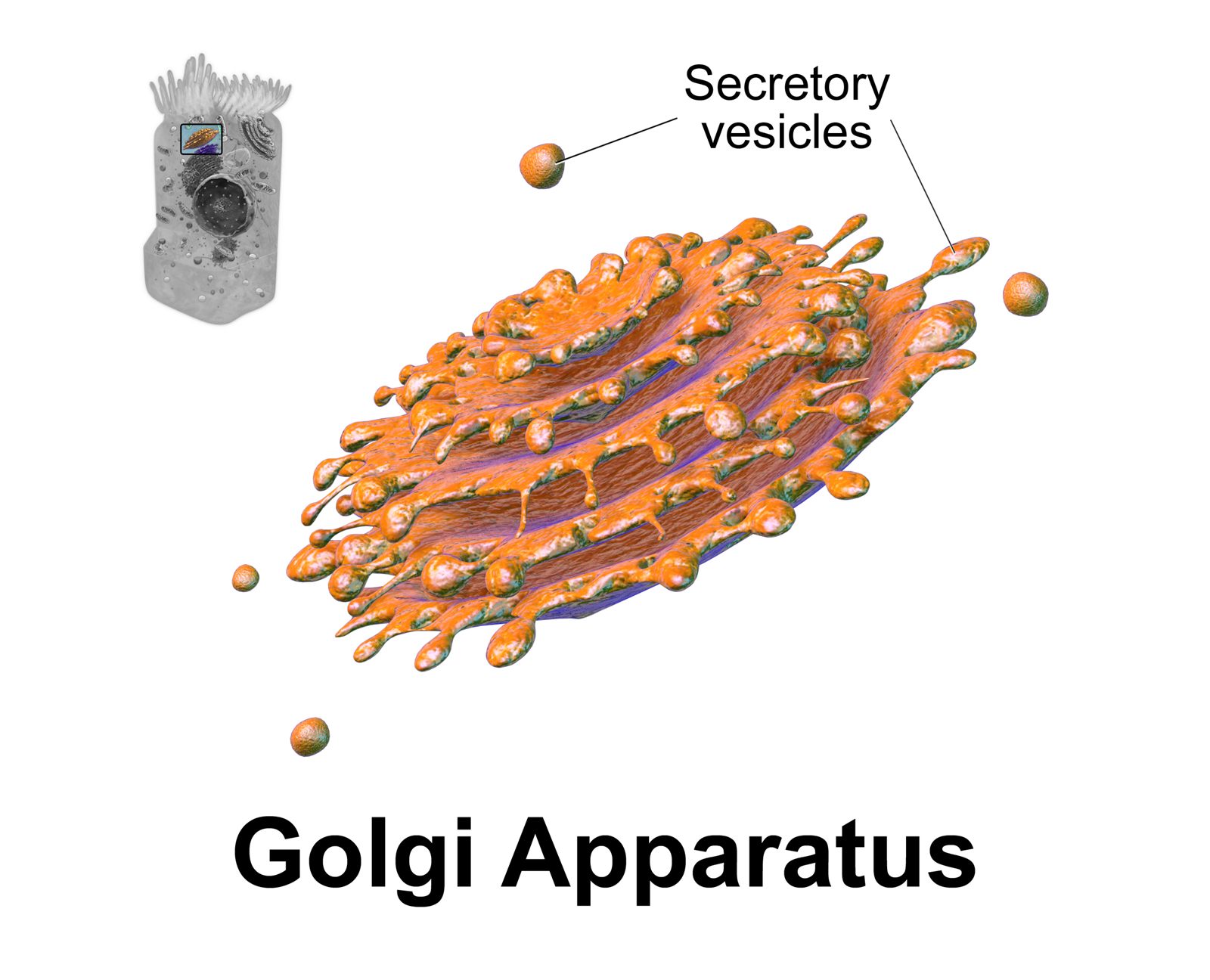
Golgi Apparatus
The Golgi apparatus (), also known as the Golgi complex, Golgi body, or simply the Golgi, is an organelle found in most eukaryotic cells. Part of the endomembrane system in the cytoplasm, it packages proteins into membrane-bound vesicles inside the cell before the vesicles are sent to their destination. It resides at the intersection of the secretory, lysosomal, and endocytic pathways. It is of particular importance in processing proteins for secretion, containing a set of glycosylation enzymes that attach various sugar monomers to proteins as the proteins move through the apparatus. It was identified in 1897 by the Italian scientist Camillo Golgi and was named after him in 1898. Discovery Owing to its large size and distinctive structure, the Golgi apparatus was one of the first organelles to be discovered and observed in detail. It was discovered in 1898 by Italian physician Camillo Golgi during an investigation of the nervous system. After first observing it under his ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteolysis
Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called proteases, but may also occur by intra-molecular digestion. Proteolysis in organisms serves many purposes; for example, digestive enzymes break down proteins in food to provide amino acids for the organism, while proteolytic processing of a polypeptide chain after its synthesis may be necessary for the production of an active protein. It is also important in the regulation of some physiological and cellular processes including apoptosis, as well as preventing the accumulation of unwanted or misfolded proteins in cells. Consequently, abnormality in the regulation of proteolysis can cause disease. Proteolysis can also be used as an analytical tool for studying proteins in the laboratory, and it may also be used in industry, for example in food proc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Structural Motif
In a polymer, chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common Biomolecular structure#Tertiary structure, three-dimensional structure that appears in a variety of different, evolutionarily unrelated molecules. A structural motif does not have to be associated with a sequence motif; it can be represented by different and completely unrelated sequences in different proteins or RNA. In nucleic acids Depending upon the sequence and other conditions, nucleic acids can form a variety of structural motifs which is thought to have biological significance. ;Stem-loop: Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Domain Of Unknown Function
A domain of unknown function (DUF) is a protein domain that has no characterised function. These families have been collected together in the Pfam database using the prefix DUF followed by a number, with examples being DUF2992 and DUF1220. As of 2019, there are almost 4,000 DUF families within the Pfam database representing over 22% of known families. Some DUFs are not named using the nomenclature due to popular usage but are nevertheless DUFs. The DUF designation is tentative, and such families tend to be renamed to a more specific name (or merged to an existing domain) after a function is identified. History The DUF naming scheme was introduced by Chris Ponting, through the addition of DUF1 and DUF2 to the SMART (database), SMART database. These two domains were found to be widely distributed in bacterial signaling proteins. Subsequently, the functions of these domains were identified and they have since been renamed as the GGDEF domain and EAL domain respectively. Characterisati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Motifs And Domains For C18orf63
Motif may refer to: General concepts * Motif (chess composition), an element of a move in the consideration of its purpose * Motif (folkloristics), a recurring element that creates recognizable patterns in folklore and folk-art traditions * Motif (music), a salient recurring fragment or succession of notes * Motif (narrative), any recurring element in a story that has symbolic significance or the reason behind actions * Motif (textile arts), a recurring element or fragment that, when joined together, creates a larger work * Motif (visual arts), a repeated theme or pattern Biochemistry * Sequence motif, a sequence pattern of nucleotides in a DNA sequence or amino acids in a protein * Short linear motif, a stretch of protein sequence that mediates protein–protein interaction * Structural motif, a pattern in a protein structure formed by the spatial arrangement of amino acids Other uses * ''Motif'' (2019 film), 2019 Malaysian Malay-language crime drama film * ''Motif'' (al ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Sheet
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, notably Alzheimer's disease. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. A refined versi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Turn
β turns (also β-bends, tight turns, reverse turns, Venkatachalam turns) are the most common form of turns—a type of non-regular secondary structure in proteins that cause a change in direction of the polypeptide chain. They are very common motifs in proteins and polypeptides. Each consists of four amino acid residues (labelled ''i'', ''i+1'', ''i+2'' and ''i+3''). They can be defined in two ways: # By the possession of an intra-main-chain hydrogen bond between the CO of residue ''i'' and the NH of residue ''i+3''; # By having a distance of less than 7Å between the Cα atoms of residues ''i'' and ''i+3''. The hydrogen bond criterion is the one most appropriate for everyday use, partly because it gives rise to four distinct categories; the distance criterion gives rise to the same four categories but yields additional turn types. Definition Hydrogen bond criterion The hydrogen bond criterion for beta turns, applied to polypeptides whose amino acids are linked by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Image Of DUF 4708
An image is a visual representation of something. It can be two-dimensional, three-dimensional, or somehow otherwise feed into the visual system to convey information. An image can be an artifact, such as a photograph or other two-dimensional picture, that resembles a subject. In the context of signal processing, an image is a distributed amplitude of color(s). In optics, the term “image” may refer specifically to a 2D image. An image does not have to use the entire visual system to be a visual representation. A popular example of this is of a greyscale image, which uses the visual system's sensitivity to brightness across all wavelengths, without taking into account different colors. A black and white visual representation of something is still an image, even though it does not make full use of the visual system's capabilities. Images are typically still, but in some cases can be moving or animated. Characteristics Images may be two or three-dimensional, such as a pho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |