|
Biosensing
A biosensor is an analytical device, used for the detection of a chemical substance, that combines a biological component with a physicochemical detector. The ''sensitive biological element'', e.g. tissue, microorganisms, organelles, cell receptors, enzymes, antibodies, nucleic acids, etc., is a biologically derived material or biomimetic component that interacts with, binds with, or recognizes the analyte under study. The biologically sensitive elements can also be created by biological engineering. The ''transducer'' or the ''detector element'', which transforms one signal into another one, works in a physicochemical way: optical, piezoelectric, electrochemical, electrochemiluminescence etc., resulting from the interaction of the analyte with the biological element, to easily measure and quantify. The biosensor reader device connects with the associated electronics or signal processors that are primarily responsible for the display of the results in a user-friendly way. Thi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biosensors Used For Screening Combinatorial DNA Libraries
A biosensor is an analytical device, used for the detection of a chemical substance, that combines a biological component with a physical chemistry, physicochemical detector. The ''sensitive biological element'', e.g. tissue, microorganisms, organelles, cell receptors, enzymes, antibody, antibodies, nucleic acids, etc., is a biologically derived material or biomimetic component that interacts with, binds with, or recognizes the analyte under study. The biologically sensitive elements can also be created by biological engineering. The Biotransducer, ''transducer'' or the ''detector element'', which transforms one signal into another one, works in a physicochemical way: optical, piezoelectric, electrochemical, electrochemiluminescence etc., resulting from the interaction of the analyte with the biological element, to easily measure and quantify. The biosensor reader device connects with the associated electronics or signal processors that are primarily responsible for the display ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biotransducer
A biotransducer is the recognition-transduction component of a biosensor system. It consists of two intimately coupled parts; a bio-recognition layer and a physicochemical transducer, which acting together converts a biochemical signal to an electronic or optical signal. The bio-recognition layer typically contains an enzyme or another binding protein such as antibody. However, oligonucleotide sequences, sub-cellular fragments such as organelles (e.g. mitochondria) and receptor carrying fragments (e.g. cell wall), single whole cells, small numbers of cells on synthetic scaffolds, or thin slices of animal or plant tissues, may also comprise the bio-recognition layer. It gives the biosensor selectivity and specificity. The physicochemical transducer is typically in intimate and controlled contact with the recognition layer. As a result of the presence and biochemical action of the analyte (target of interest), a physico-chemical change is produced within the biorecognition layer t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Physical Chemistry
Physical chemistry is the study of macroscopic and microscopic phenomena in chemical systems in terms of the principles, practices, and concepts of physics such as motion, energy, force, time, thermodynamics, quantum chemistry, statistical mechanics, analytical dynamics and chemical equilibria. Physical chemistry, in contrast to chemical physics, is predominantly (but not always) a supra-molecular science, as the majority of the principles on which it was founded relate to the bulk rather than the molecular or atomic structure alone (for example, chemical equilibrium and colloids). Some of the relationships that physical chemistry strives to resolve include the effects of: # Intermolecular forces that act upon the physical properties of materials ( plasticity, tensile strength, surface tension in liquids). # Reaction kinetics on the rate of a reaction. # The identity of ions and the electrical conductivity of materials. # Surface science and electrochemistry of cel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antigen
In immunology, an antigen (Ag) is a molecule or molecular structure or any foreign particulate matter or a pollen grain that can bind to a specific antibody or T-cell receptor. The presence of antigens in the body may trigger an immune response. The term ''antigen'' originally referred to a substance that is an antibody generator. Antigens can be proteins, peptides (amino acid chains), polysaccharides (chains of monosaccharides/simple sugars), lipids, or nucleic acids. Antigens are recognized by antigen receptors, including antibodies and T-cell receptors. Diverse antigen receptors are made by cells of the immune system so that each cell has a specificity for a single antigen. Upon exposure to an antigen, only the lymphocytes that recognize that antigen are activated and expanded, a process known as clonal selection. In most cases, an antibody can only react to and bind one specific antigen; in some instances, however, antibodies may cross-reactivity, cross-react and bind more tha ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Binding Constant
The binding constant, or affinity constant/association constant, is a special case of the equilibrium constant ''K'', and is the inverse of the dissociation constant. It is associated with the binding and unbinding reaction of receptor (R) and ligand (L) molecules, which is formalized as: :R + L RL The reaction is characterized by the on-rate constant ''k''on and the off-rate constant ''k''off, which have units of M−1 s−1 and s−1, respectively. In equilibrium, the forward binding transition R + L → RL should be balanced by the backward unbinding transition RL → R + L. That is, :k_\,[]\,[] = k_\,[], where [R], [L] and [RL] represent the concentration of unbound free receptors, the concentration of unbound free ligand and the concentration of receptor-ligand complexes. The binding constant ''K''a is defined by :K_ = = . An often considered quantity is the dissociation constant ''K''d ≡ , which has the unit of concentration, despite the fact that str ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MRNA Display
mRNA display is a display technique used for ''in vitro'' protein, and/or peptide evolution to create molecules that can bind to a desired target. The process results in translated peptides or proteins that are associated with their mRNA progenitor via a puromycin linkage. The complex then binds to an immobilized target in a selection step (affinity chromatography). The mRNA-protein fusions that bind well are then reverse transcribed to cDNA and their sequence amplified via a polymerase chain reaction. The result is a nucleotide sequence that encodes a peptide with high affinity for the molecule of interest. Puromycin is an analogue of the 3’ end of a tyrosyl-tRNA with a part of its structure mimics a molecule of adenosine, and the other part mimics a molecule of tyrosine. Compared to the cleavable ester bond in a tyrosyl-tRNA, puromycin has a non-hydrolysable amide bond. As a result, puromycin interferes with translation, and causes premature release of translation products. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Yeast Display
Yeast display (or yeast surface display) is a protein engineering technique that uses the expression of recombinant proteins incorporated into the cell wall of yeast for isolating and engineering antibodies. Development The yeast display technique was first published by the laboratory of Professor K. Dane Wittrup and Eric T. Boder. The technology was sold to Abbott Laboratories in 2001. How it works A protein of interest is displayed as a fusion to the Aga2p protein on the surface of yeast. The Aga2p protein is naturally used by yeast to mediate cell–cell contacts during yeast cell mating. As such, display of a protein via Aga2p projects the protein away from the cell surface, minimizing potential interactions with other molecules on the yeast cell wall. The use of magnetic separation and flow cytometry in conjunction with a yeast display library is a highly effective method to isolate high affinity protein ligands against nearly any receptor through directed evolution. A ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosome Display
Ribosome display is a technique used to perform '' in vitro'' protein evolution to create proteins that can bind to a desired ligand. The process results in translated proteins that are associated with their mRNA progenitor which is used, as a complex, to bind to an immobilized ligand in a selection step. The mRNA-protein hybrids that bind well are then reverse transcribed to cDNA and their sequence amplified via PCR. The result is a nucleotide sequence that can be used to create tightly binding proteins. Ribosome display process Ribosome display begins with a native library of DNA sequences coding for polypeptides. Each sequence is transcribed, and then translated ''in vitro'' into polypeptide. However, the DNA library coding for a particular library of binding proteins is genetically fused to a spacer sequence lacking a stop codon before its end. The lack of a stop codon prevents release factors from binding and triggering the disassembly of the translational complex. So, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phage Display
Phage display is a laboratory technique for the study of protein–protein, protein–peptide, and protein– DNA interactions that uses bacteriophages (viruses that infect bacteria) to connect proteins with the genetic information that encodes them. In this technique, a gene encoding a protein of interest is inserted into a phage coat protein gene, causing the phage to "display" the protein on its outside while containing the gene for the protein on its inside, resulting in a connection between genotype and phenotype. These displaying phages can then be screened against other proteins, peptides or DNA sequences, in order to detect interaction between the displayed protein and those other molecules. In this way, large libraries of proteins can be screened and amplified in a process called ''in vitro'' selection, which is analogous to natural selection. The most common bacteriophages used in phage display are M13 and fd filamentous phage, though T4, T7, and λ phage have ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
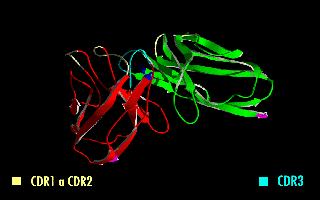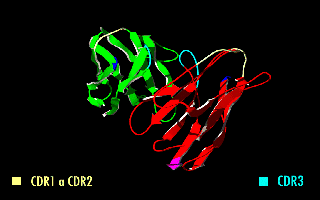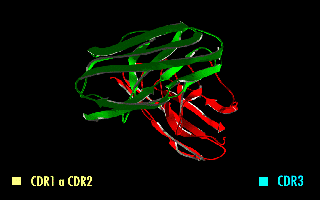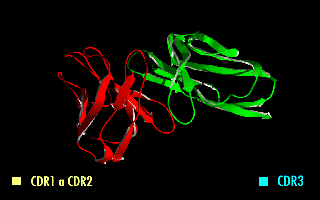
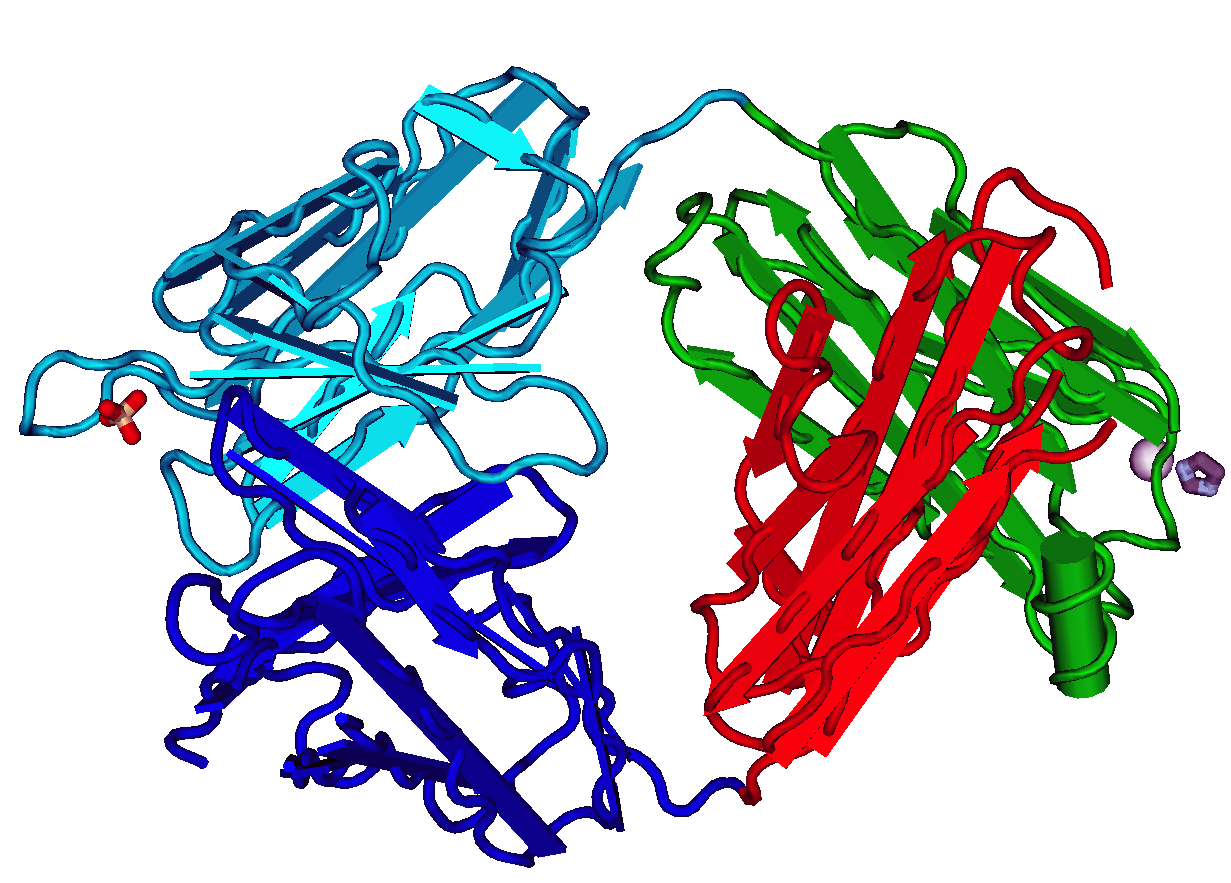
ScFv
A single-chain variable fragment (scFv) is not actually a fragment of an antibody, but instead is a fusion protein of the variable regions of the heavy (VH) and light chains (VL) of immunoglobulins, connected with a short linker peptide of ten to about 25 amino acids. The linker is usually rich in glycine for flexibility, as well as serine or threonine for solubility, and can either connect the N-terminus of the VH with the C-terminus of the VL, or ''vice versa''. This protein retains the specificity of the original immunoglobulin, despite removal of the constant regions and the introduction of the linker. The image to the right shows how this modification usually leaves the specificity unaltered. These molecules were created to facilitate phage display, where it is highly convenient to express the antigen-binding domain as a single peptide. As an alternative, scFv can be created directly from subcloned heavy and light chains derived from a hybridoma. ScFvs have many uses, e.g., ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fragment Variable
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein used by the immune system to identify and neutralize foreign objects such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the pathogen, called an antigen. Each tip of the "Y" of an antibody contains a paratope (analogous to a lock) that is specific for one particular epitope (analogous to a key) on an antigen, allowing these two structures to bind together with precision. Using this binding mechanism, an antibody can ''tag'' a microbe or an infected cell for attack by other parts of the immune system, or can neutralize it directly (for example, by blocking a part of a virus that is essential for its invasion). To allow the immune system to recognize millions of different antigens, the antigen-binding sites at both tips of the antibody come in an equally wide variety. In contrast, the remainder of the antibody is relatively constant. It only occurs in a few vari ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fragment Antigen-binding
The fragment antigen-binding region (Fab region) is a region on an antibody that binds to antigens. It is composed of one constant and one variable domain of each of the heavy and the light chain. The variable domain contains the paratope (the antigen-binding site), comprising a set of complementarity-determining regions, at the amino terminal end of the monomer. Each arm of the Y thus binds an epitope on the antigen. Preparation In an experimental setting, Fc and Fab fragments can be generated in the laboratory. The enzyme papain can be used to cleave an immunoglobulin monomer into two Fab fragments and an Fc fragment. Conversely, the enzyme pepsin cleaves below the hinge region, so the result instead is a F(ab')2 fragment and a pFc' fragment. Recently another enzyme for generation of F(ab')2 has been commercially available. The enzyme IdeS (Immunoglobulin degrading enzyme from ''Streptococcus pyogenes'', trade name FabRICATOR) cleaves IgG in a sequence specific manner at ne ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |