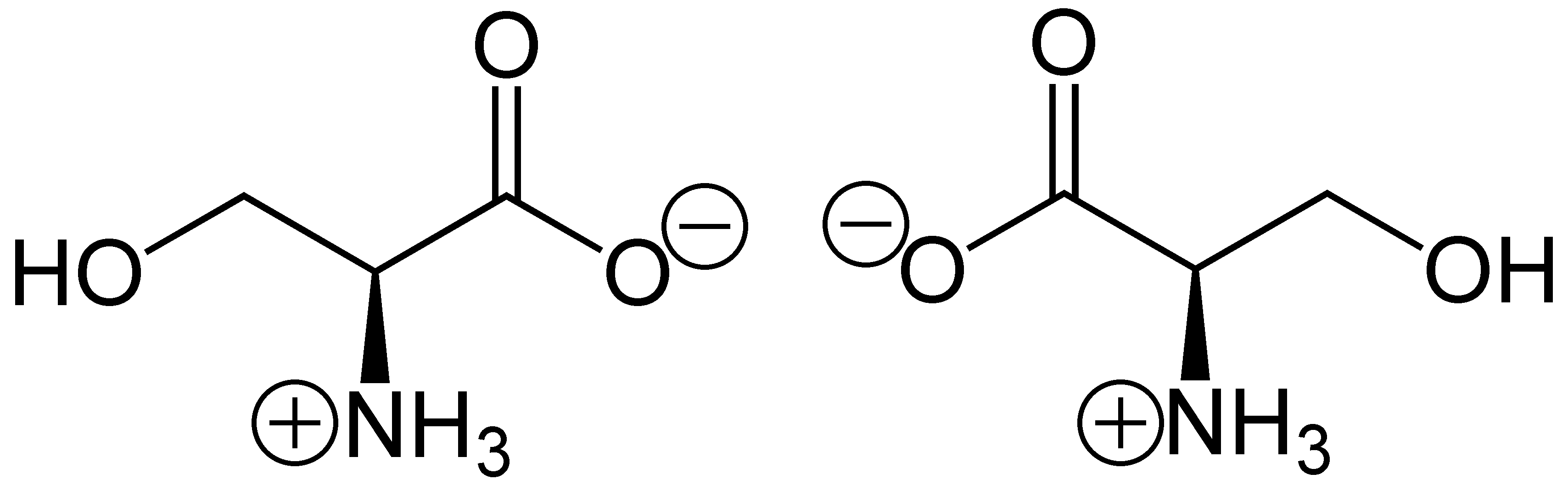|
AEBSF
AEBSF or 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride is a water-soluble, irreversible serine protease inhibitor with a molecular weight of 239.5 Da. It inhibits proteases like chymotrypsin, kallikrein, plasmin, thrombin, and trypsin. The specificity is similar to the inhibitor PMSF, nevertheless AEBSF is more stable at low pH values. Typical usage is 0.1 - 1.0 mM. Mechanism of action Both AEBSF and PMSF are sulfonyl fluorides and are sulfonylating agents. Sulfonyl fluorides act by reacting with the hydroxy group of the active site serine residue to form a sulfonyl enzyme derivative. This derivative may be stable for long periods of time except at high pH. Use in cholesterol regulation studies AEBSF is extensively used in studies aiming to describe cholesterol regulatory genes due to its potent ability to inhibit Site-1-protease (S1P). This serine protease, located in the Golgi apparatus, is responsible for activating the sterol regulatory element-binding protei ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine Protease
Serine proteases (or serine endopeptidases) are enzymes that cleave peptide bonds in proteins. Serine serves as the nucleophilic amino acid at the (enzyme's) active site. They are found ubiquitously in both eukaryotes and prokaryotes. Serine proteases fall into two broad categories based on their structure: chymotrypsin-like (trypsin-like) or subtilisin-like. Classification The MEROPS protease classification system counts 16 superfamilies (as of 2013) each containing many families. Each superfamily uses the catalytic triad or dyad in a different protein fold and so represent convergent evolution of the catalytic mechanism. The majority belong to the S1 family of the PA clan (superfamily) of proteases. For superfamilies, P: superfamily, containing a mixture of nucleophile class families, S: purely serine proteases. superfamily. Within each superfamily, families are designated by their catalytic nucleophile, (S: serine proteases). Substrate specificity Serine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PMSF
In biochemistry, phenylmethylsulfonyl fluoride (PMSF) is a serine protease inhibitor (serine hydrolase inactivator) commonly used in the preparation of cell lysates. PMSF does not inactivate all serine protease Serine proteases (or serine endopeptidases) are enzymes that cleave peptide bonds in proteins. Serine serves as the nucleophilic amino acid at the (enzyme's) active site. They are found ubiquitously in both eukaryotes and prokaryotes. Seri ...s. The effective concentration of PMSF is between 0.1 - 1 mM. The half-life is short in aqueous solutions (110 min at pH 7, 55 min at pH 7.5, and 35 min at pH 8, all at 25 °C). Stock solutions are usually made up in anhydrous ethanol, isopropanol, or corn oil and diluted immediately before use. PMSF binds specifically to the active site serine residue in serine hydrolases. It does not bind to any other serine residues in the protein. This is a result of the hyperactivity of that serine residue caused by the specific env ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine Protease Inhibitors
Serpins are a superfamily of proteins with similar structures that were first identified for their protease inhibition activity and are found in all kingdoms of life. The acronym serpin was originally coined because the first serpins to be identified act on chymotrypsin-like serine proteases (serine protease inhibitors). They are notable for their unusual mechanism of action, in which they irreversibly inhibit their target protease by undergoing a large conformational change to disrupt the target's active site. This contrasts with the more common competitive mechanism for protease inhibitors that bind to and block access to the protease active site. Protease inhibition by serpins controls an array of biological processes, including coagulation and inflammation, and consequently these proteins are the target of medical research. Their unique conformational change also makes them of interest to the structural biology and protein folding research communities. The conformatio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MEROPS
MEROPS is an online database for peptidases (also known as proteases, proteinases and proteolytic enzymes) and their inhibitors. The classification scheme for peptidases was published by Rawlings & Barrett in 1993, and that for protein inhibitors by Rawlings ''et al.'' in 2004.Rawlings, N.D., Tolle, D.P. & Barrett, A.J. (2004) "Evolutionary families of peptidase inhibitors." ''Biochem J'' 378, 705-716. The most recent version, MEROPS 12.3, was released in September 2020. Overview The classification is based on similarities at the tertiary and primary structural levels. Comparisons are restricted to that part of the sequence directly involved in the reaction, which in the case of a peptidase must include the active site, and for a protein inhibitor the reactive site. The classification is hierarchical: sequences are assembled into families, and families are assembled into clans. Each peptidase, family, and clan has a unique identifier. Classification Family The families of pe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine
Serine (symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − form under biological conditions), and a side chain consisting of a hydroxymethyl group, classifying it as a polar amino acid. It can be synthesized in the human body under normal physiological circumstances, making it a nonessential amino acid. It is encoded by the codons UCU, UCC, UCA, UCG, AGU and AGC. Occurrence This compound is one of the naturally occurring proteinogenic amino acids. Only the L-stereoisomer appears naturally in proteins. It is not essential to the human diet, since it is synthesized in the body from other metabolites, including glycine. Serine was first obtained from silk protein, a particularly rich source, in 1865 by Emil Cramer. Its name is derived from the Latin for silk, ''sericum''. Serine's structure was estab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Active Site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate (binding site) and residues that catalyse a reaction of that substrate (catalytic site). Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes. Each active site is evolved to be optimised to bind a particular substrate and catalyse a particular reaction, resulting in high specificity. This specificity is determined by the arrangement of amino acids within the active site and the structure of the substrates. Sometimes enzymes also need to bind with some cofactors to fulfil their function. The active si ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydroxyl
In chemistry, a hydroxy or hydroxyl group is a functional group with the chemical formula and composed of one oxygen atom covalently bonded to one hydrogen atom. In organic chemistry, alcohols and carboxylic acids contain one or more hydroxy groups. Both the negatively charged anion , called hydroxide, and the neutral radical , known as the hydroxyl radical, consist of an unbonded hydroxy group. According to IUPAC definitions, the term ''hydroxyl'' refers to the hydroxyl radical () only, while the functional group is called a ''hydroxy group''. Properties Water, alcohols, carboxylic acids, and many other hydroxy-containing compounds can be readily deprotonated due to a large difference between the electronegativity of oxygen (3.5) and that of hydrogen (2.1). Hydroxy-containing compounds engage in intermolecular hydrogen bonding increasing the electrostatic attraction between molecules and thus to higher boiling and melting points than found for compounds that lack this f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sulfonyl Fluoride
In inorganic chemistry, sulfonyl halide groups occur when a sulfonyl () functional group is singly bonded to a halogen atom. They have the general formula , where X is a halogen. The stability of sulfonyl halides decreases in the order fluorides > chlorides > bromides > iodides, all four types being well known. The sulfonyl chlorides and fluorides are of dominant importance in this series. Structure Sulfonyl halides have tetrahedral sulfur centres attached to two oxygen atoms, an organic radical, and a halide. In a representative example, methanesulfonyl chloride, the S=O, S−C, and S−Cl bond distances are respectively 142.4, 176.3, and 204.6 pm. Sulfonyl chlorides Sulfonic acid chlorides, or sulfonyl chlorides, are a sulfonyl halide with the general formula . Production Arylsulfonyl chlorides are made industrially in a two-step, one-pot reaction from an arene (in this case, benzene) and chlorosulfuric acid: :C6H6 + HOSO2Cl -> C6H5SO3H + HCl :C6H5SO3H + HOSO2Cl -> C6 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Trypsin
Trypsin is an enzyme in the first section of the small intestine that starts the digestion of protein molecules by cutting these long chains of amino acids into smaller pieces. It is a serine protease from the PA clan superfamily, found in the digestive system of many vertebrates, where it hydrolyzes proteins. Trypsin is formed in the small intestine when its proenzyme form, the trypsinogen produced by the pancreas, is activated. Trypsin cuts peptide chains mainly at the carboxyl side of the amino acids lysine or arginine. It is used for numerous biotechnological processes. The process is commonly referred to as trypsin proteolysis or trypsinization, and proteins that have been digested/treated with trypsin are said to have been trypsinized. Trypsin was discovered in 1876 by Wilhelm Kühne and was named from the Ancient Greek word for rubbing since it was first isolated by rubbing the pancreas with glycerin. Function In the duodenum, trypsin catalyzes the hydrolysis of pept ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme Inhibitor
An enzyme inhibitor is a molecule that binds to an enzyme and blocks its activity. Enzymes are proteins that speed up chemical reactions necessary for life, in which substrate molecules are converted into products. An enzyme facilitates a specific chemical reaction by binding the substrate to its active site, a specialized area on the enzyme that accelerates the most difficult step of the reaction. An enzyme inhibitor stops ("inhibits") this process, either by binding to the enzyme's active site (thus preventing the substrate itself from binding) or by binding to another site on the enzyme such that the enzyme's catalysis of the reaction is blocked. Enzyme inhibitors may bind reversibly or irreversibly. Irreversible inhibitors form a chemical bond with the enzyme such that the enzyme is inhibited until the chemical bond is broken. By contrast, reversible inhibitors bind non-covalently and may spontaneously leave the enzyme, allowing the enzyme to resume its function. Reve ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Thrombin
Thrombin (, ''fibrinogenase'', ''thrombase'', ''thrombofort'', ''topical'', ''thrombin-C'', ''tropostasin'', ''activated blood-coagulation factor II'', ''blood-coagulation factor IIa'', ''factor IIa'', ''E thrombin'', ''beta-thrombin'', ''gamma-thrombin'') is a serine protease, an enzyme that, in humans, is encoded by the ''F2'' gene. Prothrombin (coagulation factor II) is proteolytically cleaved to form thrombin in the clotting process. Thrombin in turn acts as a serine protease that converts soluble fibrinogen into insoluble strands of fibrin, as well as catalyzing many other coagulation-related reactions. History After the description of fibrinogen and fibrin, Alexander Schmidt hypothesised the existence of an enzyme that converts fibrinogen into fibrin in 1872. Prothrombin was discovered by Pekelharing in 1894. Physiology Synthesis Thrombin is produced by the enzymatic cleavage of two sites on prothrombin by activated Factor X (Xa). The activity of factor Xa is greatly ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plasmin
Plasmin is an important enzyme () present in blood that degrades many blood plasma proteins, including fibrin clots. The degradation of fibrin is termed fibrinolysis. In humans, the plasmin protein (in the zymogen form of plasminogen) is encoded by the ''PLG'' gene. Function Plasmin is a serine protease that acts to dissolve fibrin blood clots. Apart from fibrinolysis, plasmin proteolyses proteins in various other systems: It activates collagenases, some mediators of the complement system, and weakens the wall of the Graafian follicle, leading to ovulation. Plasmin is also integrally involved in inflammation. It cleaves fibrin, fibronectin, thrombospondin, laminin, and von Willebrand factor. Plasmin, like trypsin, belongs to the family of serine proteases. Plasmin is released as a zymogen called plasminogen (PLG) from the liver into the systemic circulation. Two major glycoforms of plasminogen are present in humans - type I plasminogen contains two glycosylation moieties ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |