Histone code on:
[Wikipedia]
[Google]
[Amazon]
The histone code is a
* H2BK5ac
Cellsignal.com Histone Modifications with function and attached references
Histone Code Overview sheet
Histone Modification Guide
{{DEFAULTSORT:Histone Code Epigenetics de:Histonmodifikation#Histon Code
hypothesis
A hypothesis (: hypotheses) is a proposed explanation for a phenomenon. A scientific hypothesis must be based on observations and make a testable and reproducible prediction about reality, in a process beginning with an educated guess o ...
that the transcription of genetic information encoded in DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
is in part regulated by chemical modifications (known as ''histone marks'') to histone proteins, primarily on their unstructured ends. Together with similar modifications such as DNA methylation it is part of the epigenetic code. Histones associate with DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
to form nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome ...
s, which themselves bundle to form chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important r ...
fibers, which in turn make up the more familiar chromosome
A chromosome is a package of DNA containing part or all of the genetic material of an organism. In most chromosomes, the very long thin DNA fibers are coated with nucleosome-forming packaging proteins; in eukaryotic cells, the most import ...
. Histones are globular proteins with a flexible N-terminus (taken to be the tail) that protrudes from the nucleosome. Many of the histone tail modifications correlate very well to chromatin structure and both histone modification state and chromatin structure correlate well to gene expression levels. The critical concept of the histone code hypothesis is that the histone modifications serve to recruit other proteins by specific recognition of the modified histone via protein domain
In molecular biology, a protein domain is a region of a protein's Peptide, polypeptide chain that is self-stabilizing and that Protein folding, folds independently from the rest. Each domain forms a compact folded Protein tertiary structure, thre ...
s specialized for such purposes, rather than through simply stabilizing or destabilizing the interaction between histone and the underlying DNA. These recruited proteins then act to alter chromatin structure actively or to promote transcription.
For details of gene expression regulation by histone modifications see table below.
The hypothesis
The hypothesis is thatchromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important r ...
-DNA interactions are guided by combinations of histone modifications. While it is accepted that modifications (such as methylation, acetylation, ADP-ribosylation, ubiquitination, citrullination, SUMO
is a form of competitive full-contact wrestling where a ''rikishi'' (wrestler) attempts to force his opponent out of a circular ring (''dohyō'') or into touching the ground with any body part other than the soles of his feet (usually by th ...
-ylation and phosphorylation
In biochemistry, phosphorylation is described as the "transfer of a phosphate group" from a donor to an acceptor. A common phosphorylating agent (phosphate donor) is ATP and a common family of acceptor are alcohols:
:
This equation can be writ ...
) to histone tails alter chromatin structure, a complete understanding of the precise mechanisms by which these alterations to histone tails influence DNA-histone interactions remains elusive. However, some specific examples have been worked out in detail. For example, phosphorylation of serine residues 10 and 28 on histone H3
Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal end, N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'b ...
is a marker for chromosomal condensation. Similarly, the combination of phosphorylation of serine residue 10 and acetylation of a lysine residue 14 on histone H3 is a tell-tale sign of active transcription.

Modifications
Well characterized modifications to histones include: * Methylation: Both lysine and arginine residues are known to be methylated. Methylated lysines are the best understood marks of the histone code, as specific methylated lysine match well with gene expression states. Methylation of lysines H3K4 and H3K36 is correlated with transcriptional activation while demethylation of H3K4 is correlated with silencing of the genomic region. Methylation of lysines H3K9 and H3K27 is correlated with transcriptional repression. Particularly, H3K9me3 is highly correlated with constitutive heterochromatin. Methylation of histone lysine also has a role inDNA repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is cons ...
. For instance, H3K36me3 is required for homologous recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in Cell (biology), cellular organi ...
al repair of DNA double-strand breaks, and H4K20me2 facilitates repair of such breaks by non-homologous end joining.
* Acetylation—by HAT
A hat is a Headgear, head covering which is worn for various reasons, including protection against weather conditions, ceremonial reasons such as university graduation, religious reasons, safety, or as a fashion accessory. Hats which incorpor ...
(histone acetyl transferase); deacetylation—by HDAC (histone deacetylase): Acetylation tends to define the 'openness' of chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important r ...
as acetylated histones cannot pack as well together as deacetylated histones.
* Phosphorylation
In biochemistry, phosphorylation is described as the "transfer of a phosphate group" from a donor to an acceptor. A common phosphorylating agent (phosphate donor) is ATP and a common family of acceptor are alcohols:
:
This equation can be writ ...
* Ubiquitination
* SUMOylation
However, there are many more histone modifications, and sensitive mass spectrometry
Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a ''mass spectrum'', a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is used ...
approaches have recently greatly expanded the catalog.
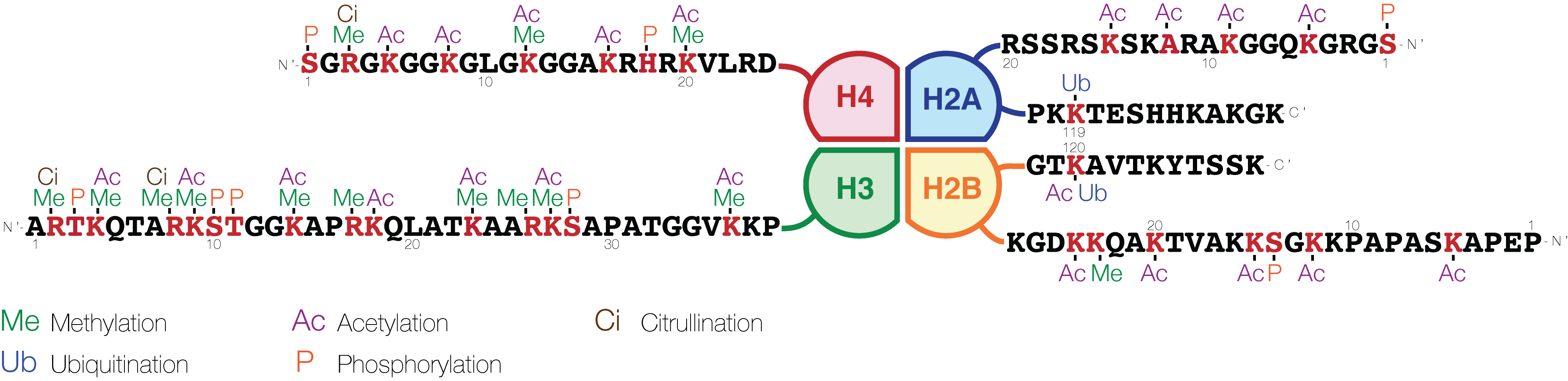
A very basic summary of the histone code for gene expression status is given below (histone nomenclature is described here):
Histone H3
Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal end, N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'b ...
* H3K4me1 - primed enhancers
* H3K4me3 is enriched in transcriptionally active promoters.
* H3K9me2 -repression
* H3K9me3 is found in constitutively repressed genes.
* H3K27me3 is found in facultatively repressed genes.
* H3K36me
* H3K36me2
* H3K36me3 is found in actively transcribed gene bodies.
* H3K79me2
* H3K9ac is found in actively transcribed promoters.
* H3K14ac is found in actively transcribed promoters.
* H3K23ac
* H3K27ac distinguishes active enhancers from poised enhancers.
* H3K36ac
* H3K56ac is a proxy for de novo histone assembly.
* H3K122ac is enriched in poised promoters and also found in a different type of putative enhancer that lacks H3K27ac.
Histone H4
Histone H4 is one of the five main histone proteins involved in the structure of chromatin in eukaryote, eukaryotic cells. Featuring a main globular domain and a long N-terminus, N-terminal tail, H4 is involved with the structure of the nucleo ...
* H4K5ac
* H4K8ac
* H4K12ac
* H4K16ac
* H4K20me
* H4K91ac
Complexity
Unlike this simplified model, any real histone code has the potential to be massively complex; each of the four standard histones can be simultaneously modified at multiple different sites with multiple different modifications. To give an idea of this complexity,histone H3
Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal end, N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'b ...
contains nineteen lysines known to be methylated—each can be un-, mono-, di- or tri-methylated. If modifications are independent, this allows a potential 419 or 280 billion different lysine methylation patterns, far more than the maximum number of histones in a human genome (6.4 Gb / ~150 bp = ~44 million histones if they are very tightly packed). And this does not include lysine acetylation (known for H3 at nine residues), arginine methylation (known for H3 at three residues) or threonine/serine/tyrosine phosphorylation (known for H3 at eight residues), not to mention modifications of other histones.
Every nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome ...
in a cell can therefore have a different set of modifications, raising the question of whether common patterns of histone modifications exist. A study of about 40 histone modifications across human gene promoters found over 4000 different combinations used, over 3000 occurring at only a single promoter. However, patterns were discovered including a set of 17 histone modifications that are present together at over 3000 genes. Mass spectrometry
Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a ''mass spectrum'', a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is used ...
-based top-down proteomics
Top-down proteomics is a method of protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, ...
has provided more insight into these patterns by being able to discriminate single molecule co-occurrence from co-localization in the genome or on the same nucleosome. A variety of approaches have been used to delve into detailed biochemical mechanisms that demonstrate the importance of interplay between histone modifications. Thus, specific patterns of histone modifications are more common than others. These patterns are functionally important but they are intricate and challenging to study. We currently have the best biochemical understanding of the importance of a relatively small number of discrete modifications and a few combinations.
Structural determinants of histone recognition by readers, writers, and erasers of the histone code are revealed by a growing body of experimental data.
See also
* Histone * Histone-modifying enzymesReferences
External links
Cellsignal.com Histone Modifications with function and attached references
Histone Code Overview sheet
Histone Modification Guide
{{DEFAULTSORT:Histone Code Epigenetics de:Histonmodifikation#Histon Code