|
QpAdm
ADMIXTOOLS (or AdmixTools) is a software package that is primarily used for analyzing admixture in population genetics. The original version was developed as a set of standalone C programs by Nick Patterson and colleagues and published in 2012. A reimplemented version, ADMIXTOOLS 2, was developed as an R package by Robert Maier and colleagues and published in 2023. Most ADMIXTOOLS programs are based on fitting demographic models to f-statistics, which are calculated from population allele frequencies. qpGraph qpGraph is a software program that is part of the ADMIXTOOLS software package developed by Patterson et al. (2012). qpGraph evaluates graph-based models of population relationships with genetic admixture. It estimates likelihoods of graphs with a fixed topology, while adjusting graph parameters to fit observed f-statistics. ADMIXTOOLS 2 adds functionality for finding optimized graph topologies, similar to programs like Treemix and OrientAGraph. Other tools Related stat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetics Software
This list of phylogenetics software is a compilation of computational phylogenetics software used to produce phylogenetic trees. Such tools are commonly used in comparative genomics, cladistics, and bioinformatics. Methods for estimating phylogenies include neighbor-joining, maximum parsimony (also simply referred to as parsimony), unweighted pair group method with arithmetic mean (UPGMA), Bayesian phylogenetic inference, maximum likelihood, and distance matrix methods. List See also * List of phylogenetic tree visualization software References External links *Complete list oInstitut Pasteurphylogeny webserversExPASyList of phylogenetics programs *A verof phylogenetic tools (reconstruction, visualization, ''etc.'') *Anothelist of evolutionary genetics software*A list ophylogenetic softwareprovided by the Zoological Research Museum A. Koenig *MicrobeTrace available at https://github.com/CDCgov/MicrobeTrace/wiki {{DEFAULTSORT:List Of Phylogenetics Software Geneti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CP/NNLS
CP/NNLS, standing for "ChromoPainter (CP) non-negative least squares (NNLS)" is a statistical method used in genetics. "ChromoPainter" is the name of a tool for finding haplotypes in sequence data, in which each individual is "painted" as a combination of all other sequences. It is used in Principal Components Analysis (PCA) to create data summaries, or dating admixture events. Non-negative least squares (NNLS) is a kind of regression analysis, which aims at finding the best possible correlation between a large set of dependent variables. It here used within the functionalities of ChromoPainter. CP/NNLS is often used in conjunction with qpAdm ADMIXTOOLS (or AdmixTools) is a software package that is primarily used for analyzing admixture in population genetics. The original version was developed as a set of standalone C programs by Nick Patterson and colleagues and published in 2012. .... References {{reflist Least squares ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Likelihood Function
A likelihood function (often simply called the likelihood) measures how well a statistical model explains observed data by calculating the probability of seeing that data under different parameter values of the model. It is constructed from the joint probability distribution of the random variable that (presumably) generated the observations. When evaluated on the actual data points, it becomes a function solely of the model parameters. In maximum likelihood estimation, the argument that maximizes the likelihood function serves as a point estimate for the unknown parameter, while the Fisher information (often approximated by the likelihood's Hessian matrix at the maximum) gives an indication of the estimate's precision. In contrast, in Bayesian statistics, the estimate of interest is the ''converse'' of the likelihood, the so-called posterior probability of the parameter given the observed data, which is calculated via Bayes' rule. Definition The likelihood function, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GitHub
GitHub () is a Proprietary software, proprietary developer platform that allows developers to create, store, manage, and share their code. It uses Git to provide distributed version control and GitHub itself provides access control, bug tracking system, bug tracking, software feature requests, task management, continuous integration, and wikis for every project. Headquartered in California, GitHub, Inc. has been a subsidiary of Microsoft since 2018. It is commonly used to host open source software development projects. GitHub reported having over 100 million developers and more than 420 million Repository (version control), repositories, including at least 28 million public repositories. It is the world's largest source code host Over five billion developer contributions were made to more than 500 million open source projects in 2024. About Founding The development of the GitHub platform began on October 19, 2005. The site was launched in April 2008 by Tom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dendroscope
Dendroscope is an interactive computer software program written in Java for viewing Phylogenetic trees. This program is designed to view trees of all sizes and is very useful for creating figures. Dendroscope can be used for a variety of analyses of molecular data sets but is particularly designed for metagenomics or analyses of uncultured environmental samples. It was developed by Daniel Huson and his colleagues at the University of Tübingen in Germany, who also created SplitsTree. See also *List of phylogenetic tree visualization software *SplitsTree *MEGAN Megan is a Welsh feminine given name, originally a diminutive form of Margaret. Margaret is from the Greek μαργαρίτης (''margarítēs''), Latin ''margarīta'', "pearl". Megan is one of the most popular Welsh-language names for women in ... References External links Dendroscope homepage hosted at the University of Washington Phylogenetics software {{science-software-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SplitsTree
SplitsTree is a freeware program for inferring phylogenetic trees, phylogenetic networks, or, more generally, split graphs, from various types of data such as a sequence alignment, a distance matrix or a set of trees. Software SplitsTree implements published methods such as split (phylogenetics), split decomposition, neighbor-net, consensus networks, super networks methods or methods for computing Hybrid (biology), hybridization or simple genetic recombination, recombination networks. It uses the Nexus file, NEXUS file format. The splits graph is defined using a special data block (SPLITS block). See also *Phylogenetic tree viewers *Dendroscope *MEGAN References {{Reflist External links SplitsTree homepage(New Website for informations about SplitsTree)for the latest version (4.15) and manual (June 2019), hosted by thDepartment of Computer Scienceat the University of Tübingen, Eberhard Karls University TübingenAlgorithms in Bioinformatics Daniel Huson's working group deve ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Clade
In biology, a clade (), also known as a Monophyly, monophyletic group or natural group, is a group of organisms that is composed of a common ancestor and all of its descendants. Clades are the fundamental unit of cladistics, a modern approach to taxonomy adopted by most biological fields. The common ancestor may be an individual, a population, or a species (extinct or Extant taxon, extant). Clades are nested, one in another, as each branch in turn splits into smaller branches. These splits reflect evolutionary history as populations diverged and evolved independently. Clades are termed ''monophyletic'' (Greek: "one clan") groups. Over the last few decades, the cladistic approach has revolutionized biological classification and revealed surprising evolutionary relationships among organisms. Increasingly, taxonomists try to avoid naming Taxon, taxa that are not clades; that is, taxa that are not Monophyly, monophyletic. Some of the relationships between organisms that the molecul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nick Patterson (scientist)
Nicholas James Patterson (born 9 June 1947) is a mathematician working as a staff scientist at the Broad Institute with notable contributions to the area of computational genomics. His work has appeared in scientific journals such as ''Nature'', ''Science'' and ''Nature Genetics''. His research has brought a better understanding of early human migrations. He is among the group of scientists who have sequenced the Neanderthal genome in 2010. This was followed by the sequencing of a much higher quality Neanderthal genome, where the subject was from the Altai Mountains, in 2014. These studies have uncovered some unexpected facts about the interbreeding between archaic and modern humans. Biography Patterson was an only child who grew up in the Bayswater section of central London. Patterson received his B.A. and Ph.D. in mathematics at Cambridge University. His doctoral advisor was John G. Thompson. He initially worked for the British code-breaking agency GCHQ and the Center ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
David Reich (geneticist)
David Emil Reich (born July 14, 1974) is an American geneticist known for the industrialization of research into the population genetics of ancient humans, including their migrations and the mixing of populations, discovered by analysis of genome-wide patterns of mutations. He is professor in the department of genetics at the Harvard Medical School, and an associate of the Broad Institute. Reich was highlighted as one of Nature's 10 for his contributions to science in 2015. He received the Dan David Prize in 2017, the NAS Award in Molecular Biology, the Wiley Prize, and the Darwin–Wallace Medal in 2019. In 2021 he was awarded the Massry Prize. He is the director of the David Reich Lab. Early life and education Reich grew up as part of a Jewish family in Washington, D.C. His parents are novelist Tova Reich (sister of Rabbi Avi Weiss) and Walter Reich, a professor at George Washington University, who served as the first director of the United States Holocaust Memoria ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
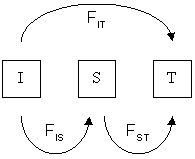
F-statistics
In population genetics, ''F''-statistics (also known as fixation indices) describe the statistically expected level of heterozygosity in a population; more specifically the expected degree of (usually) a reduction in heterozygosity when compared to Hardy–Weinberg expectation. ''F''-statistics can also be thought of as a measure of the correlation between genes drawn at different levels of a (hierarchically) subdivided population. This correlation is influenced by several evolution Evolution is the change in the heritable Phenotypic trait, characteristics of biological populations over successive generations. It occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, re ...ary processes, such as genetic drift, founder effect, population bottleneck, bottleneck, genetic hitchhiking, meiotic drive, mutation, gene flow, inbreeding, natural selection, or the Wahlund effect, but it was originally designed to measure the amount ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |