F-statistics on:
[Wikipedia]
[Google]
[Amazon]
In
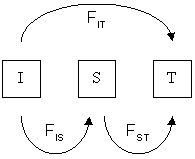 Consider a population that has a population structure of two levels; one from the individual (I) to the subpopulation (S) and one from the subpopulation to the total (T). Then the total , known here as , can be partitioned into and :
:
This may be further partitioned for population substructure, and it expands according to the rules of binomial expansion, so that for ''I'' partitions:
:
Consider a population that has a population structure of two levels; one from the individual (I) to the subpopulation (S) and one from the subpopulation to the total (T). Then the total , known here as , can be partitioned into and :
:
This may be further partitioned for population substructure, and it expands according to the rules of binomial expansion, so that for ''I'' partitions:
:
Shane's Simple Guide to F-Statistics
Analyzing the genetic structure of populations
* ttps://web.archive.org/web/20160303172553/http://helix.mcmaster.ca/brent/node10.html IAM based F-statistics
F-statistics for Population Genetics Eco-Tool
Population Structure (slides)
{{DEFAULTSORT:F-Statistics Population genetics
population genetics
Population genetics is a subfield of genetics that deals with genetic differences within and among populations, and is a part of evolutionary biology. Studies in this branch of biology examine such phenomena as Adaptation (biology), adaptation, s ...
, ''F''-statistics (also known as fixation indices) describe the statistically expected level of heterozygosity
Zygosity (the noun, zygote, is from the Greek "yoked," from "yoke") () is the degree to which both copies of a chromosome or gene have the same genetic sequence. In other words, it is the degree of similarity of the alleles in an organism.
Mos ...
in a population; more specifically the expected degree of (usually) a reduction in heterozygosity when compared to Hardy–Weinberg expectation.
''F''-statistics can also be thought of as a measure of the correlation between genes drawn at different levels of a (hierarchically) subdivided population. This correlation is influenced by several evolution
Evolution is the change in the heritable Phenotypic trait, characteristics of biological populations over successive generations. It occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, re ...
ary processes, such as genetic drift
Genetic drift, also known as random genetic drift, allelic drift or the Wright effect, is the change in the Allele frequency, frequency of an existing gene variant (allele) in a population due to random chance.
Genetic drift may cause gene va ...
, founder effect
In population genetics, the founder effect is the loss of genetic variation that occurs when a new population is established by a very small number of individuals from a larger population. It was first fully outlined by Ernst Mayr in 1942, us ...
, bottleneck
Bottleneck may refer to:
* the narrowed portion (neck) of a bottle
Science and technology
* Bottleneck (engineering), where the performance of an entire system is limited by a single component
* Bottleneck (network), in a communication network
* ...
, genetic hitchhiking
Genetic hitchhiking, also called genetic draft or the hitchhiking effect, is when an allele changes frequency not because it itself is under natural selection, but because it is near another gene that is undergoing a selective sweep and that is ...
, meiotic drive, mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, ...
, gene flow
In population genetics, gene flow (also known as migration and allele flow) is the transfer of genetic variation, genetic material from one population to another. If the rate of gene flow is high enough, then two populations will have equivalent ...
, inbreeding
Inbreeding is the production of offspring from the mating or breeding of individuals or organisms that are closely genetic distance, related genetically. By analogy, the term is used in human reproduction, but more commonly refers to the genet ...
, natural selection
Natural selection is the differential survival and reproduction of individuals due to differences in phenotype. It is a key mechanism of evolution, the change in the Heredity, heritable traits characteristic of a population over generation ...
, or the Wahlund effect, but it was originally designed to measure the amount of allelic fixation owing to genetic drift
Genetic drift, also known as random genetic drift, allelic drift or the Wright effect, is the change in the Allele frequency, frequency of an existing gene variant (allele) in a population due to random chance.
Genetic drift may cause gene va ...
.
The concept of ''F''-statistics was developed during the 1920s by the American geneticist Sewall Wright
Sewall Green Wright ForMemRS
HonFRSE (December 21, 1889March 3, 1988) was an American geneticist known for his influential work on evolutionary theory and also for his work on path analysis. He was a founder of population genetics alongside ...
, who was interested in inbreeding in cattle
Cattle (''Bos taurus'') are large, domesticated, bovid ungulates widely kept as livestock. They are prominent modern members of the subfamily Bovinae and the most widespread species of the genus '' Bos''. Mature female cattle are calle ...
. However, because complete dominance causes the phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology (physical form and structure), its developmental processes, its biochemical and physiological propert ...
s of homozygote
Zygosity (the noun, zygote, is from the Greek "yoked," from "yoke") () is the degree to which both copies of a chromosome or gene have the same genetic sequence. In other words, it is the degree of similarity of the alleles in an organism.
Mos ...
dominants and heterozygotes to be the same, it was not until the advent of molecular genetics
Molecular genetics is a branch of biology that addresses how differences in the structures or expression of DNA molecules manifests as variation among organisms. Molecular genetics often applies an "investigative approach" to determine the st ...
from the 1960s onwards that heterozygosity in populations could be measured.
''F'' can be used to define effective population size
The effective population size (''N'e'') is the size of an idealised population that would experience the same rate of genetic drift as the real population. Idealised populations are those following simple one- locus models that comply with ass ...
.
Definitions and equations
The measures FIS, FST, and FIT are related to the amounts of heterozygosity at various levels of population structure. Together, they are called ''F''-statistics, and are derived from ''F'', theinbreeding coefficient
The coefficient of relationship is a measure of the degree of consanguinity (or biological relationship) between two individuals. The term coefficient of relationship was defined by Sewall Wright in 1922, and was derived from his definition of ...
. In a simple two-allele system with inbreeding, the genotypic frequencies are:
:
The value for is found by solving the equation for using heterozygotes in the above inbred population. This becomes one minus the observed frequency of heterozygotes in a population divided by the expected frequency of heterozygotes at Hardy–Weinberg equilibrium:
:
where the expected frequency at Hardy–Weinberg equilibrium is given by
:
where and are the allele frequencies of and , respectively. It is also the probability that at any locus, two alleles from a random individual of the population are identical by descent.
For example, consider the data from E.B. Ford (1971) on a single population of the scarlet tiger moth:
From this, the allele frequencies can be calculated, and the expectation of derived :
:
:
:
The different F-statistics look at different levels of population structure. FIT is the inbreeding coefficient of an individual (I) relative to the total (T) population, as above; FIS is the inbreeding coefficient of an individual (I) relative to the subpopulation (S), using the above for subpopulations and averaging them; and FST is the effect of subpopulations (S) compared to the total population (T), and is calculated by solving the equation:
:
as shown in the next section.
Partition due to population structure
 Consider a population that has a population structure of two levels; one from the individual (I) to the subpopulation (S) and one from the subpopulation to the total (T). Then the total , known here as , can be partitioned into and :
:
This may be further partitioned for population substructure, and it expands according to the rules of binomial expansion, so that for ''I'' partitions:
:
Consider a population that has a population structure of two levels; one from the individual (I) to the subpopulation (S) and one from the subpopulation to the total (T). Then the total , known here as , can be partitioned into and :
:
This may be further partitioned for population substructure, and it expands according to the rules of binomial expansion, so that for ''I'' partitions:
:
Fixation index
A reformulation of the definition of would be the ratio of the average number of differences between pairs of chromosomes sampled within diploid individuals with the average number obtained when sampling chromosomes randomly from the population (excluding the grouping per individual). One can modify this definition and consider a grouping per sub-population instead of per individual. Population geneticists have used that idea to measure the degree of structure in a population. Unfortunately, there is a large number of definitions for , causing some confusion in the scientific literature. A common definition is the following: : where the variance of is computed across sub-populations and is the expected frequency of heterozygotes.Fixation index in human populations
It is well established that the genetic diversity among human populations is low, although the distribution of the genetic diversity was only roughly estimated. Early studies argued that 85–90% of the genetic variation is found within individuals residing in the same populations within continents (intra-continental populations) and only an additional 10–15% is found between populations of different continents (continental populations). Later studies based on hundreds of thousandssingle-nucleotide polymorphism
In genetics and bioinformatics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a ...
(SNPs) suggested that the genetic diversity between continental populations is even smaller and accounts for 3 to 7% A later study based on three million SNPs found that 12% of the genetic variation is found between continental populations and only 1% within them. Most of these studies have used the ''F''''ST'' statistics or closely related statistics.
See also
* Malecot's method of coancestry *Heterozygosity
Zygosity (the noun, zygote, is from the Greek "yoked," from "yoke") () is the degree to which both copies of a chromosome or gene have the same genetic sequence. In other words, it is the degree of similarity of the alleles in an organism.
Mos ...
* Hardy-Weinberg principle
* Wahlund effect
* QST-FST analyses
* Coefficient of inbreeding
*Coefficient of relationship
The coefficient of relationship is a measure of the degree of consanguinity (or biological relationship) between two individuals. The term coefficient of relationship was defined by Sewall Wright in 1922, and was derived from his definition of th ...
*Fixation index
The fixation index (FST) is a measure of population differentiation due to genetic structure. It is frequently estimated from Polymorphism (biology), genetic polymorphism data, such as single-nucleotide polymorphisms (SNP) or Microsatellite (genet ...
References
External links
Shane's Simple Guide to F-Statistics
Analyzing the genetic structure of populations
* ttps://web.archive.org/web/20160303172553/http://helix.mcmaster.ca/brent/node10.html IAM based F-statistics
F-statistics for Population Genetics Eco-Tool
Population Structure (slides)
{{DEFAULTSORT:F-Statistics Population genetics