|
Mir-143
In molecular biology mir-143 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms. mir–143 is highly conserved in vertebrates. mir-143 is thought be involved in cardiac morphogenesis but has also been implicated in cancer. Genomic location mir– 143 is located on chromosome 5 position 33 in the human genome. mir-143 is located very close to mir-145 in the genome and it is speculated that they are transcribed as a bicistronic unit. Their co-transcription means they are frequently studied together in the same cellular pathways and diseases. Expression mir–143 is a direct transcriptional target of the serum response factor, myocardin and nkx2-5. mir-143 expression is also thought to be controlled epigenetically through heart beat. Targets These are known genetic targets for mir–143 and its effect on them: * Klf4 – Promotes transcription. * ELK1 – Promotes transcription. * ADD3 – Represses ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MicroRNA
Micro ribonucleic acid (microRNA, miRNA, μRNA) are small, single-stranded, non-coding RNA molecules containing 21–23 nucleotides. Found in plants, animals, and even some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRNAs base-pair to complementary sequences in messenger RNA (mRNA) molecules, then silence said mRNA molecules by one or more of the following processes: * Cleaving the mRNA strand into two pieces. * Destabilizing the mRNA by shortening its poly(A) tail. * Reducing translation of the mRNA into proteins. In cells of humans and other animals, miRNAs primarily act by destabilizing the mRNA. miRNAs resemble the small interfering RNAs (siRNAs) of the RNA interference (RNAi) pathway, except miRNAs derive from regions of RNA transcripts that fold back on themselves to form short stem-loops (hairpins), whereas siRNAs derive from longer regions of double-stranded RNA. The human genome may encode ov ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ADD3
Gamma-adducin is a protein that in humans is encoded by the ''ADD3'' gene. Adducins are heteromeric proteins composed of different subunits referred to as adducin alpha, beta and gamma. The three subunits are encoded by distinct genes and belong to a family of membrane skeletal proteins involved in the assembly of spectrin-actin network in erythrocytes and at sites of cell-cell contact in epithelial tissues. While adducins alpha and gamma are ubiquitously expressed, the expression of adducin beta is restricted to brain and hematopoietic tissues. Adducin, originally purified from human erythrocytes, was found to be a heterodimer of adducins alpha and beta. Polymorphisms resulting in amino acid substitutions in these two subunits have been associated with the regulation of blood pressure in an animal model of hypertension. Heterodimers consisting of alpha and gamma subunits have also been described. Structurally, each subunit is composed of two distinct domains. The amino-te ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metastasis
Metastasis is a pathogenic agent's spreading from an initial or primary site to a different or secondary site within the host's body; the term is typically used when referring to metastasis by a cancerous tumor. The newly pathological sites, then, are metastases (mets). It is generally distinguished from cancer invasion, which is the direct extension and penetration by cancer cells into neighboring tissues. Cancer occurs after cells are genetically altered to proliferate rapidly and indefinitely. This uncontrolled proliferation by mitosis produces a primary tumor, primary tumour heterogeneity, heterogeneic tumour. The cells which constitute the tumor eventually undergo metaplasia, followed by dysplasia then anaplasia, resulting in a Malignancy, malignant phenotype. This malignancy allows for invasion into the circulation, followed by invasion to a second site for tumorigenesis. Some cancer cells, known as circulating tumor cells (CTCs), are able to penetrate the walls of lymp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UBE2E3
Ubiquitin-conjugating enzyme E2 E3 is a protein that in humans is encoded by the ''UBE2E3'' gene. The modification of proteins with ubiquitin is an important cellular mechanism for targeting abnormal or short-lived proteins for degradation. Ubiquitination involves at least three classes of enzymes: ubiquitin-activating enzymes, or E1s, ubiquitin-conjugating enzymes, or E2s, and ubiquitin-protein ligases, or E3s. This gene encodes a member of the E2 ubiquitin-conjugating enzyme family. The encoded protein shares 100% sequence identity with the mouse and rat counterparts, which indicates that this enzyme is highly conserved in eukaryotes. Two alternatively spliced Alternative splicing, alternative RNA splicing, or differential splicing, is an alternative RNA splicing, splicing process during gene expression that allows a single gene to produce different splice variants. For example, some exons of a gene ma ... transcript variants encoding the same protein have been found for th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
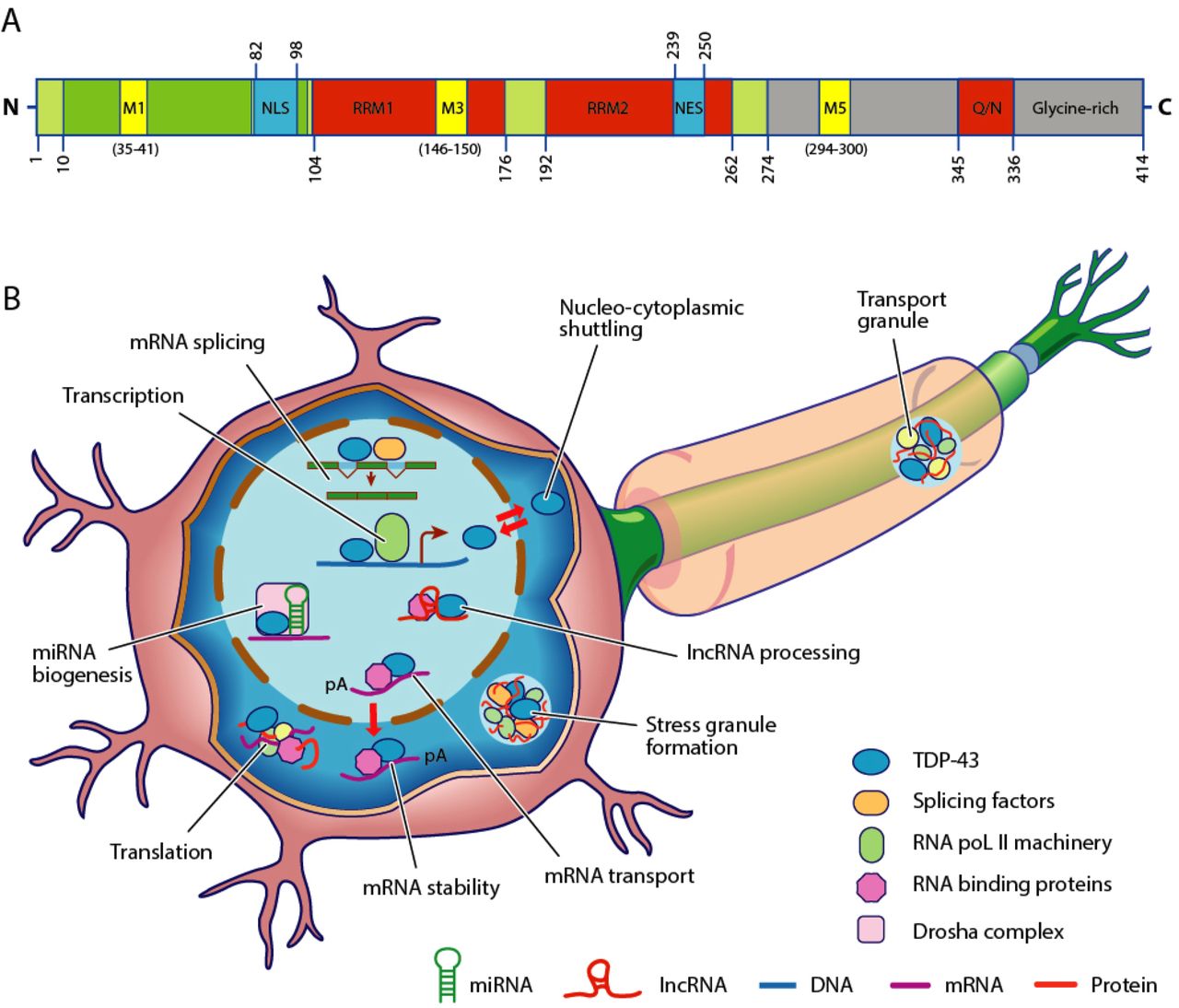
TARDBP
Transactive response DNA binding protein 43 kDa (TAR DNA-binding protein 43 or TDP-43) is a protein that in humans is encoded by the ''TARDBP'' gene. Structure TDP-43 is 414 amino acid residues long. It consists of four domains: an N-terminal domain spanning residues 1–76 (NTD) with a well-defined fold that has been shown to form a dimer or oligomer; two highly conserved folded RNA recognition motifs spanning residues 106–176 (RRM1) and 191–259 (RRM2), respectively, required to bind target RNA and DNA; an unstructured C-terminal domain encompassing residues 274–414 (CTD), which contains a glycine-rich region, is involved in protein-protein interactions, and harbors most of the mutations associated with familial amyotrophic lateral sclerosis. The entire protein devoid of large solubilising tags has been purified. The full-length protein is a dimer. The dimer is formed due to a self-interaction between two NTD domains, where the dimerisation can be propagated to fo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MAP3K7
Mitogen-activated protein kinase kinase kinase 7 (MAP3K7), also known as TAK1, is an enzyme that in humans is encoded by the ''MAP3K7'' gene. Structure TAK1 is an evolutionarily conserved kinase in the MAP3 K family and clusters with the tyrosine-like and sterile kinase families. The protein structure of TAK1 contains an N (residues 1–104)- and C (residues 111–303)-terminus connected through the hinge region (Met 104-Ser 111). The ATP binding pocket is located in the hinge region of the kinase. Additionally, TAK1 has a catalytic lysine (Lys63) in the active site. Crystal structure of TAK1-ATP have shown that ATP forms two hydrogen bonds with residues Ala 107 and Glu 105. Further hydrogen bonding is observed to Asp 175, which is the leading residue of the DFG motif. This residue is thought to interact with Lys 63 through polar interactions and is catalytically important for phosphate transfer to substrate molecules. Critical for the TAK1-TAB1 complex is a helical loop arou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ELK1
ETS Like-1 protein Elk-1 is a protein that in humans is encoded by the ELK1. Elk-1 functions as a transcription activator. It is classified as a ternary complex factor (TCF), a subclass of the ETS family, which is characterized by a common protein domain that regulates DNA binding to target sequences. Elk1 plays important roles in various contexts, including long-term memory formation, drug addiction, Alzheimer's disease, Down syndrome, breast cancer, and Major depressive disorder, depression. Structure As depicted in Figure 1, the Elk1 protein is composed of several domains. Localized in the N-terminal region, the A domain is required for the binding of Elk1 to DNA. This region also contains a nuclear localization signal (NLS) and a nuclear export signal (NES), which are responsible for nuclear import and export, respectively. The B domain allows Elk1 to bind to a dimer of its cofactor, serum response factor (SRF). Located adjacent to the B domain, the R domain is involved in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryote
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms are eukaryotes. They constitute a major group of Outline of life forms, life forms alongside the two groups of prokaryotes: the Bacteria and the Archaea. Eukaryotes represent a small minority of the number of organisms, but given their generally much larger size, their collective global biomass is much larger than that of prokaryotes. The eukaryotes emerged within the archaeal Kingdom (biology), kingdom Asgard (Archaea), Promethearchaeati and its sole phylum Promethearchaeota. This implies that there are only Two-domain system, two domains of life, Bacteria and Archaea, with eukaryotes incorporated among the Archaea. Eukaryotes first emerged during the Paleoproterozoic, likely as Flagellated cell, flagellated cells. The leading evolutiona ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mir-145 MicroRNA
In molecular biology, mir-145 microRNA is a short RNA molecule that in humans is encoded by the MIR145 gene. MicroRNAs function to regulate the expression levels of other genes by several mechanisms. Targets MicroRNAs are involved in down-regulation of a variety of target genes. Götte ''et al''. have shown that experimental over-expression of mir-145 down-regulates the junctional cell adhesion molecule JAM-A as well as the actin bundling protein fascin in breast cancer and endometriosis cells, resulting in a reduction of cell motility. Larsson ''et al''. showed that miR-145 targets the 3' UTR of the FLI1 gene, a finding that was later supported by Zhang ''et al''. Role in cancer miR-145 is hypothesised to be a tumor suppressor. miR-145 has been shown to be down-regulated in breast cancer. miR-145 is also involved in colon cancer and acute myeloid leukemia Acute myeloid leukemia (AML) is a cancer of the myeloid line of blood cells, characterized by the rapid growth of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |