|
YAP1
YAP1 (yes-associated protein 1), also known as YAP or YAP65, is a protein that acts as a transcription coregulator that promotes transcription of genes involved in cellular proliferation and suppressing apoptotic genes. YAP1 is a component in the hippo signaling pathway which regulates organ size, regeneration, and tumorigenesis. YAP1 was first identified by virtue of its ability to associate with the SH3 domain of Yes and Src protein tyrosine kinases. ''YAP1'' is a potent oncogene, which is amplified in various human cancers. Structure Cloning of the YAP1 gene facilitated the identification of a modular protein domain, known as the WW domain. Two splice isoforms of the YAP1 gene product were initially identified, named YAP1-1 and YAP1-2, which differed by the presence of an extra 38 amino acids that encoded the WW domain. Apart from the WW domain, the modular structure of YAP1 contains a proline-rich region at the very amino terminus, which is followed by a TID (TEAD tran ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
WW Domain
The WW domain, (also known as the rsp5-domain or WWP repeating motif) is a modular protein domain that mediates specific interactions with protein ligands. This domain is found in a number of unrelated signaling and structural proteins and may be repeated up to four times in some proteins. Apart from binding preferentially to proteins that are proline-rich, with particular proline-motifs, PP-P- PY, some WW domains bind to phosphoserine- phosphothreonine-containing motifs. Structure and ligands The WW domain is one of the smallest protein modules, composed of only 40 amino acids, which mediates specific protein-protein interactions with short proline-rich or proline-containing motifs. Named after the presence of two conserved tryptophans (W), which are spaced 20-22 amino acids apart within the sequence, the WW domain folds into a meandering triple-stranded beta sheet. The identification of the WW domain was facilitated by the analysis of two splice isoforms of YAP gene pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
WWTR1
WW domain-containing transcription regulator protein 1 (WWTR1), also known as Transcriptional coactivator with PDZ-binding motif (TAZ), is a protein that in humans is encoded by the ''WWTR1'' gene. WWTR1 acts as a transcriptional coregulator and has no effect on transcription alone. When in complex with transcription factor binding partners, WWTR1 helps promote gene expression in pathways associated with development, cell growth and survival, and inhibiting apoptosis. Aberrant WWTR1 function has been implicated for its role in driving cancers. WWTR1 is often referred to as TAZ due to its initial characterization with the name TAZ. However, WWTR1 (TAZ) is not to be confused with the protein tafazzin, which originally held the official gene symbol TAZ, and is now TAFAZZIN. Structure WWTR1 contains a proline rich region, TEAD binding motif, WW domain, coiled coil region, and a transactivation domain (TAD) containing the PDZ domain-binding motif. WWTR1 (TAZ) lacks a DNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
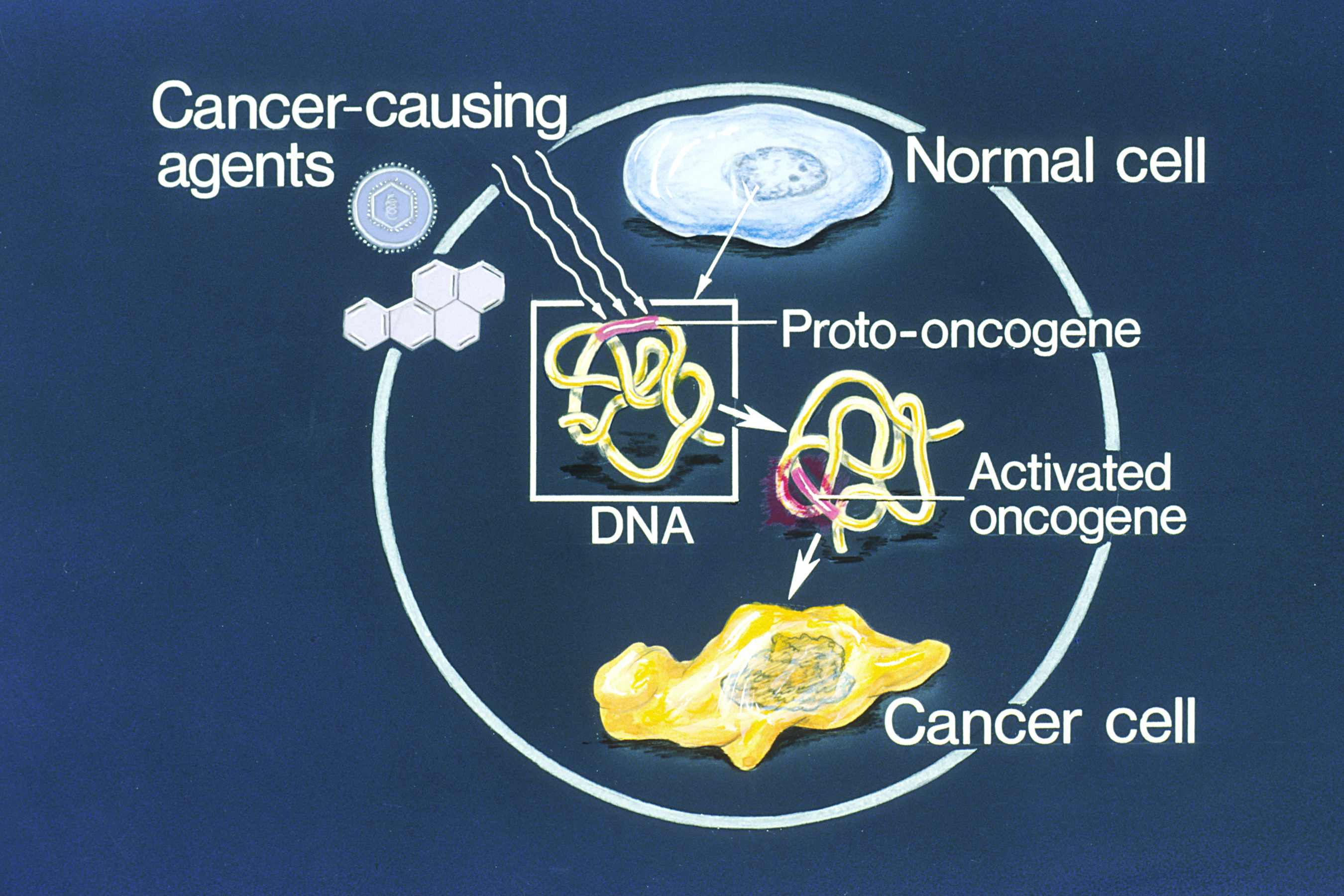
Oncogene
An oncogene is a gene that has the potential to cause cancer. In tumor cells, these genes are often mutated, or expressed at high levels.Kimball's Biology Pages. "Oncogenes" Free full text Most normal cells will undergo a programmed form of rapid cell death () when critical functions are altered and malfunctioning. Activated oncogenes can cause those cells designated for apoptosis to survive and proliferate instead. Most oncogenes began as proto-oncogenes: normal genes involved in cell growth and proliferation or inhibition of apoptosis. If, through mutation, normal genes promoting cellular growth are up-regulated (gain-of-function mutation), they will predis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
AMOT
Angiomotin (AMOT) is a protein that in humans is encoded by the ''AMOT'' gene. It belongs to the motin family of angiostatin binding proteins, which includes angiomotin, angiomotin-like 1 ( AMOTL1) and angiomotin-like 2 ( AMOTL2) characterized by coiled-coil domains at N-terminus and consensus PDZ-binding domain at the C-terminus. Angiomotin is expressed predominantly in endothelial cells of capillaries as well as angiogenic tissues such as placenta and solid tumor. Discovery Angiomotin was discovered in 2001 by screening a placenta yeast two-hybrid cDNA library for angiostatin-binding peptides, using a construct encoding the kringle domains 1-4 of angiostatin. Gene location ''AMOT'' gene is located on human chromosome X:112,021,794-112,066,354, containing 3252 nucleotides in coding sequence as 11 exons. Protein structure Two splice isoforms are known for angiomotin: p80 and p130. The alternative splicing is somewhat tissue specific. Cells expressing p130 contained more a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ErbB4
Receptor tyrosine-protein kinase erbB-4 is an enzyme that in humans is encoded by the ''ERBB4'' gene. Alternatively spliced variants that encode different protein isoforms have been described; however, not all variants have been fully characterized. Function Receptor tyrosine-protein kinase erbB-4 is a receptor tyrosine kinase that is a member of the epidermal growth factor receptor family. ERBB4 is a single-pass type I transmembrane protein with multiple furin-like cysteine rich domains, a tyrosine kinase domain, a phosphotidylinositol-3 kinase binding site and a PDZ domain binding motif. The protein binds to and is activated by neuregulins-2, -3 and -4, heparin-binding EGF-like growth factor and betacellulin. Ligand binding induces a variety of cellular responses including mitogenesis and differentiation. Multiple proteolytic events allow for the release of a cytoplasmic fragment and an extracellular fragment. Clinical significance Mutations in this gene have been assoc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Coregulator
In molecular biology and genetics, transcription coregulators are proteins that interact with transcription factors to either activate or repress the transcription of specific genes. Transcription coregulators that activate gene transcription are referred to as coactivators while those that repress are known as corepressors. The mechanism of action of transcription coregulators is to modify chromatin structure and thereby make the associated DNA more or less accessible to transcription. In humans several dozen to several hundred coregulators are known, depending on the level of confidence with which the characterisation of a protein as a coregulator can be made. One class of transcription coregulators modifies chromatin structure through covalent modification of histones. A second ATP dependent class modifies the conformation of chromatin. Histone acetyltransferases Nuclear DNA is normally tightly wrapped around histones rendering the DNA inaccessible to the general transc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SH3 Domain
The SRC Homology 3 Domain (or SH3 domain) is a small protein domain of about 60 amino acid residues. Initially, SH3 was described as a conserved sequence in the viral adaptor protein v-Crk. This domain is also present in the molecules of phospholipase and several cytoplasmic tyrosine kinases such as Abl and Src. It has also been identified in several other protein families such as: PI3 Kinase, Ras GTPase-activating protein, CDC24 and cdc25. SH3 domains are found in proteins of signaling pathways regulating the cytoskeleton, the Ras protein, and the Src kinase and many others. The SH3 proteins interact with adaptor proteins and tyrosine kinases. Interacting with tyrosine kinases, SH3 proteins usually bind far away from the active site. Approximately 300 SH3 domains are found in proteins encoded in the human genome. In addition to that, the SH3 domain was responsible for controlling protein-protein interactions in the signal transduction pathways and regulating the interac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hippo Signaling Pathway
The Hippo signaling pathway, also known as the Salvador-Warts-Hippo (SWH) pathway, is a signaling pathway that controls organ size in animals through the regulation of cell proliferation and apoptosis. The pathway takes its name from one of its key signaling components—the protein kinase Hippo (Hpo). Mutations in this gene lead to tissue overgrowth, or a "hippopotamus"-like phenotype. A fundamental question in developmental biology is how an organ knows to stop growing after reaching a particular size. Organ growth relies on several processes occurring at the cellular level, including cell division and programmed cell death (or apoptosis). The Hippo signaling pathway is involved in restraining cell proliferation and promoting apoptosis. As many cancers are marked by unchecked cell division, this signaling pathway has become increasingly significant in the study of human cancer. The Hippo pathway also has a critical role in stem cell and tissue specific progenitor cell self-ren ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proto-oncogene Tyrosine-protein Kinase Src
Proto-oncogene tyrosine-protein kinase Src, also known as proto-oncogene c-Src, or simply c-Src (cellular Src; pronounced "sarc", as it is short for sarcoma), is a non-receptor tyrosine kinase protein that in humans is encoded by the ''SRC'' gene. It belongs to a family of Src family kinases and is similar to the v-Src (viral Src) gene of Rous sarcoma virus. It includes an SH2 domain, an SH3 domain and a tyrosine kinase domain. Two transcript variants encoding the same protein have been found for this gene. c-Src phosphorylates specific tyrosine residues in other tyrosine kinases. It plays a role in the regulation of embryonic development and cell growth. An elevated level of activity of c-Src is suggested to be linked to cancer progression by promoting other signals. Mutations in c-Src could be involved in the malignant progression of colon cancer. c-Src should not be confused with Tyrosine-protein kinase CSK, CSK (C-terminal Src kinase), an enzyme that phosphorylates c-Src at it ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factors
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization ( body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Paralog
Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal (or lateral) gene transfer event (xenologs). Homology among DNA, RNA, or proteins is typically inferred from their nucleotide or amino acid sequence similarity. Significant similarity is strong evidence that two sequences are related by evolutionary changes from a common ancestral sequence. Alignments of multiple sequences are used to indicate which regions of each sequence are homologous. Identity, similarity, and conservation The term "percent homology" is often used to mean "sequence similarity”, that is the percentage of identical residues (''percent identity''), or the percentage of residues conserved with similar physicochemical properties ( ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TJP2
Tight junction protein ZO-2 is a protein that in humans is encoded by the ''TJP2'' gene. Tight junction proteins (TJPs) belong to a family of membrane-associated guanylate kinase (MAGUK) homologs that are involved in the organization of epithelial and endothelial intercellular junctions. TJPs bind to the cytoplasmic C termini of junctional transmembrane proteins and link them to the actin cytoskeleton upplied by OMIM Interactions Tight junction protein 2 has been shown to interact with tight junction protein 1, band 4.1, occludin Occludin is an enzyme ( EC 1.6) that oxidizes NADH. It was first identified in epithelial cells as a 65 kDa integral plasma-membrane protein localized at the tight junctions. Together with Claudins, and zonula occludens-1 (ZO-1), occludin has be ... and USP53. References Further reading * * * * * * * * * * * * * * * * * {{PDB Gallery, geneid=9414 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |