|
Vector NTI
Vector NTI was a commercial bioinformatics software package used by many life scientists in the early 2000s to work, among other things, with nucleic acids and proteins '' in silico''. It allowed researchers to, for example, plan a DNA cloning experiment on the computer before actually performing it in the lab. It was originally created by InforMax Inc, North Bethesda, MD in 1993 and versions in the early 2000s were well reviewed at the time. However, in 2008 it was locked and turned into a commercial software after 2008 which created problems for locked in users who were forced to buy the software to continue accessing their data on newer computers. What was previously a single software package was subsequently split into Vector NTI Express, Advanced, and Express Designer. Vector NTI was discontinued by its corporate parent Thermo Fisher at the end of 2019 and support ceased a year later. Features * create, annotate, analyse, and share DNA/ protein sequences * perform an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Life Technologies (Thermo Fisher Scientific)
Life Technologies Corporation was a biotech company founded in November 2008 through a US $6.7billion merger of Invitrogen Corporation and Applied Biosystems Inc. The joint sales of the combined companies were about $3.5 billion; they had about 9,500 employees and owned more than 3,600 licenses and patents. Company name The name "Life Technologies" was an old name from the history of Invitrogen. GIBCO (Grand Island Biological Company) had been founded around 1960 in New York; in 1983 GIBCO merged with a reagent company called Bethesda Research Laboratories and the merged company was named Life Technologies. In 2000, Invitrogen acquired Life Technologies and discontinued that name. When Invitrogen and Applied Biosystems merged, the companies revived the name. The use of the "Life Technologies" brand name is disputed. Life Technologies (India) Private Limited, a company founded in 2002, operating in this corporate name claims ownership of the brand name. The "Life Technologies" ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cloning
Cloning is the process of producing individual organisms with identical or virtually identical DNA, either by natural or artificial means. In nature, some organisms produce clones through asexual reproduction. In the field of biotechnology, cloning is the process of creating cloned organisms (copies) of cells and of DNA fragments (molecular cloning). Etymology Coined by Herbert J. Webber, the term clone derives from the Ancient Greek word (), ''twig'', which is the process whereby a new plant is created from a twig. In botany, the term ''lusus'' was used. In horticulture, the spelling ''clon'' was used until the early twentieth century; the final ''e'' came into use to indicate the vowel is a "long o" instead of a "short o". Since the term entered the popular lexicon in a more general context, the spelling ''clone'' has been used exclusively. Natural cloning Cloning is a natural form of reproduction that has allowed life forms to spread for hundreds of millions of years. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Map
A restriction map is a map of known restriction sites within a sequence of DNA. Restriction mapping requires the use of restriction enzymes. In molecular biology, restriction maps are used as a reference to engineer plasmids or other relatively short pieces of DNA, and sometimes for longer genomic DNA. There are other ways of mapping features on DNA for longer length DNA molecules, such as mapping by transduction. One approach in constructing a restriction map of a DNA molecule is to sequence the whole molecule and to run the sequence through a computer program that will find the recognition sites that are present for every restriction enzyme known. Before sequencing was automated, it would have been prohibitively expensive to sequence an entire DNA strand. To find the relative positions of restriction sites on a plasmid, a technique involving single and double restriction digests is used. Based on the sizes of the resultant DNA fragments the positions of the sites can be infer ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Open Source Bioinformatics Software
This is a list of computer software which is made for bioinformatics and released under open-source software licenses with articles in Wikipedia. See also * List of sequence alignment software * List of open-source healthcare software * List of biomedical cybernetics software * List of freeware health software * List of genetic engineering software * List of molecular graphics systems * List of systems biology modelling software * Comparison of software for molecular mechanics modeling * List of proprietary bioinformatics software References External links Free Biology Software– Free Software Directory – Free Software Foundation {{DEFAULTSORT:Open source bioinformatics software Lists of bioinformatics software * Bioinformatics Open Open or OPEN may refer to: Music * Open (band), Australian pop/rock band * The Open (band), English indie rock band * Open (Blues Image album), ''Open'' (Blues Image album), 1969 * Open (Gotthard album), ''Open'' (Gotthard album), ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Expression Vector
An expression vector, otherwise known as an expression construct, is usually a plasmid or virus designed for gene expression in cells. The vector is used to introduce a specific gene into a target cell, and can commandeer the cell's mechanism for protein synthesis to produce the protein encoded by the gene. Expression vectors are the basic tools in biotechnology for the production of proteins. The vector is engineered to contain regulatory sequences that act as enhancer and promoter regions and lead to efficient transcription of the gene carried on the expression vector. The goal of a well-designed expression vector is the efficient production of protein, and this may be achieved by the production of significant amount of stable messenger RNA, which can then be translated into protein. The expression of a protein may be tightly controlled, and the protein is only produced in significant quantity when necessary through the use of an inducer, in some systems however the protein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Computational Biology
Computational biology refers to the use of data analysis, mathematical modeling and Computer simulation, computational simulations to understand biological systems and relationships. An intersection of computer science, biology, and big data, the field also has foundations in applied mathematics, chemistry, and genetics. It differs from biological computing, a subfield of computer engineering which uses bioengineering to build computers. History Bioinformatics, the analysis of informatics processes in biological systems, began in the early 1970s. At this time, research in artificial intelligence was using network models of the human brain in order to generate new algorithms. This use of biological data pushed biological researchers to use computers to evaluate and compare large data sets in their own field. By 1982, researchers shared information via Punched card, punch cards. The amount of data grew exponentially by the end of the 1980s, requiring new computational method ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cloning Vector
A cloning vector is a small piece of DNA that can be stably maintained in an organism, and into which a foreign DNA fragment can be inserted for cloning purposes. The cloning vector may be DNA taken from a virus, the cell of a higher organism, or it may be the plasmid of a bacterium. The vector contains features that allow for the convenient insertion of a DNA fragment into the vector or its removal from the vector, for example through the presence of restriction sites. The vector and the foreign DNA may be treated with a restriction enzyme that cuts the DNA, and DNA fragments thus generated contain either blunt ends or overhangs known as sticky ends, and vector DNA and foreign DNA with compatible ends can then be joined together by molecular ligation. After a DNA fragment has been cloned into a cloning vector, it may be further subcloned into another vector designed for more specific use. There are many types of cloning vectors, but the most commonly used ones are genetical ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatics
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combines biology, chemistry, physics, computer science, information engineering, mathematics and statistics to analyze and interpret the biological data. Bioinformatics has been used for '' in silico'' analyses of biological queries using computational and statistical techniques. Bioinformatics includes biological studies that use computer programming as part of their methodology, as well as specific analysis "pipelines" that are repeatedly used, particularly in the field of genomics. Common uses of bioinformatics include the identification of candidates genes and single nucleotide polymorphisms ( SNPs). Often, such identification is made with the aim to better understand the genetic basis of disease, unique adaptations, desirable properties ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
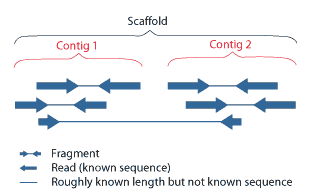
Contigs
A contig (from ''contiguous'') is a set of overlapping DNA segments that together represent a consensus region of DNA.Gregory, S. ''Contig Assembly''. Encyclopedia of Life Sciences, 2005. In bottom-up sequencing projects, a contig refers to overlapping sequence data ( reads); in top-down sequencing projects, contig refers to the overlapping clones that form a physical map of the genome that is used to guide sequencing and assembly.Dear, P. H. ''Genome Mapping''. Encyclopedia of Life Sciences, 2005. . Contigs can thus refer both to overlapping DNA sequences and to overlapping physical segments (fragments) contained in clones depending on the context. Original definition of contig In 1980, Staden wrote: ''In order to make it easier to talk about our data gained by the shotgun method of sequencing we have invented the word "contig". A contig is a set of gel readings that are related to one another by overlap of their sequences. All gel readings belong to one and only one co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromatogram
In chemical analysis, chromatography is a laboratory technique for the separation of a mixture into its components. The mixture is dissolved in a fluid solvent (gas or liquid) called the ''mobile phase'', which carries it through a system (a column, a capillary tube, a plate, or a sheet) on which a material called the ''stationary phase'' is fixed. Because the different constituents of the mixture tend to have different affinities for the stationary phase and are retained for different lengths of time depending on their interactions with its surface sites, the constituents travel at different apparent velocities in the mobile fluid, causing them to separate. The separation is based on the differential partitioning between the mobile and the stationary phases. Subtle differences in a compound's partition coefficient result in differential retention on the stationary phase and thus affect the separation. Chromatography may be preparative or analytical. The purpose of preparativ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Entrez
The Entrez (pronounced ''ɒnˈtreɪ'') Global Query Cross-Database Search System is a federated search engine, or web portal that allows users to search many discrete health sciences databases at the National Center for Biotechnology Information (NCBI) website. The NCBI is a part of the National Library of Medicine (NLM), which is itself a department of the National Institutes of Health (NIH), which in turn is a part of the United States Department of Health and Human Services. The name "Entrez" (a greeting meaning "Come in" in French) was chosen to reflect the spirit of welcoming the public to search the content available from the NLM. Entrez Global Query is an integrated search and retrieval system that provides access to all databases simultaneously with a single query string and user interface. Entrez can efficiently retrieve related sequences, structures, and references. The Entrez system can provide views of gene and protein sequences and chromosome maps. Some textbooks a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
National Center For Biotechnology Information
The National Center for Biotechnology Information (NCBI) is part of the United States National Library of Medicine (NLM), a branch of the National Institutes of Health (NIH). It is approved and funded by the government of the United States. The NCBI is located in Bethesda, Maryland, and was founded in 1988 through legislation sponsored by US Congressman Claude Pepper. The NCBI houses a series of databases relevant to biotechnology and biomedicine and is an important resource for bioinformatics tools and services. Major databases include GenBank for DNA sequences and PubMed, a bibliographic database for biomedical literature. Other databases include the NCBI Epigenomics database. All these databases are available online through the Entrez search engine. NCBI was directed by David Lipman, one of the original authors of the BLAST sequence alignment program and a widely respected figure in bioinformatics. GenBank NCBI had responsibility for making available the GenBank DNA seq ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
.jpg)