|
Tetrahydromethanopterin
Tetrahydromethanopterin (THMPT, ) is a coenzyme in methanogenesis. It is the carrier of the C1 group as it is reduced to the methyl level, before transferring to the coenzyme M. Tetrahydrosarcinapterin (THSPT, ) is a modified form of THMPT, wherein a glutamyl group linked to the 2-hydroxy glutaric acid terminus. THMPT is the main platform for C1 transformations N-Formyl methanofuran donates the C1 group to the N5 site of the pterin to give the formyl- THMPT. The formyl group subsequently condenses intramolecularly to give methenyl-, which is then reduced to methylene-THMPT by 5,10-methenyl- hydrogenase with as the electron donor. Methylene- MPT is subsequently converted, using coenzyme F420 as the electron source, to methyl-THMPT, catalyzed by F420-dependent methylene-THMPT reductase. Methyl-THMPT is the methyl donor to coenzyme M, a conversion mediated by methyl-THMPT: coenzyme M methyltransferase. Comparison with tetrahydrofolic acid THMPT is related to the bette ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
5,10-methylenetetrahydromethanopterin Reductase
In enzymology, a 5,10-methylenetetrahydromethanopterin reductase () is an enzyme that catalyzes the chemical reaction :5-methyltetrahydromethanopterin + coenzyme F420 \rightleftharpoons 5,10-methylenetetrahydromethanopterin + reduced coenzyme F420 Thus, the two substrates of this enzyme are 5-methyltetrahydromethanopterin and coenzyme F420, whereas its two products are 5,10-methylenetetrahydromethanopterin and reduced coenzyme F420. This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-NH group of donors with other acceptors. The systematic name of this enzyme class is 5-methyltetrahydromethanopterin:coenzyme-F420 oxidoreductase. Other names in common use include 5,10-methylenetetrahydromethanopterin cyclohydrolase, N5,N10-methylenetetrahydromethanopterin reductase, methylene-H4MPT reductase, coenzyme F420-dependent N5,N10-methenyltetrahydromethanopterin, reductase, and N5,N10-methylenetetrahydromethanopterin:coenzyme-F420 oxidoreductase. T ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrogenase
A hydrogenase is an enzyme that Catalysis, catalyses the reversible Redox, oxidation of molecular hydrogen (H2), as shown below: Hydrogen oxidation () is coupled to the reduction of electron acceptors such as oxygen, nitrate, Ferric, ferric ion, sulfate, carbon dioxide (), and fumarate. On the other hand, proton reduction () is coupled to the oxidation of electron donors such as ferredoxin (FNR), and serves to dispose excess electrons in cells (essential in pyruvate fermentation). Both low-molecular weight compounds and proteins such as FNRs, cytochrome ''c''3, and cytochrome ''c''6 can act as physiological electron donors or acceptors for hydrogenases. Structural classification It has been estimated that 99% of all organisms utilize hydrogen, H2. Most of these species are microbes and their ability to use H2 as a metabolite arises from the expression of metalloenzymes known as hydrogenases. Hydrogenases are sub-classified into three different types based on the active site ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coenzyme F420
Coenzyme F420 is a family of coenzymes involved in redox reactions in a number of bacteria and archaea. It is derived from coenzyme FO (7,8-didemethyl-8-hydroxy-5-deazariboflavin) and differs by having a oligoglutamyl tail attached via a 2-phospho-L-lactate bridge. F420 is so named because it is a flavin derivative with an absorption maximum at 420 nm. F420 was originally discovered in methanogenic archaea and in Actinomycetota (especially in ''Mycobacterium''). It is now known to be used also by Cyanobacteria and by soil Proteobacteria, Chloroflexi and Firmicutes. Eukaryotes including the fruit fly ''Drosophila melanogaster'' and the algae ''Ostreococcus tauri'' also use Coenzyme FO. F420 is structurally similar to FMN, but catalytically it is similar to NAD and NADP: it has low redox potential and always transfer a hydride. As a result, it is not only a versatile cofactor in biochemical reactions, but also being eyed for potential as an industrial catalyst. Similar t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methanofuran
Methanofurans (MFRs) are a family of chemical compounds found in methanogenic archaea. These species feature a 2-aminomethylfuran linked to phenoxy group. At least three different end groups are recognized: R = tricarboxyheptanoyl (methanofuran), glutamyl-glutamyl (methanofuran b), tricarboxy-2-hydroxyheptanoyl (methanofuran c, see picture). Formylation of MFR Methanofuran converts to formylmethanofuran in an early stage of methanogenesis. The enzyme formylmethanofuran dehydrogenase ( EC: 1.2.99.5) formylates methanofuran using , the primary C1 source in methanogenesis. Deformylation of MFR The enzyme formylmethanofuran:tetrahydromethanopterin formyltransferase catalyzes the transfer of the formyl group from formylmethanofuran to N5 on tetrahydromethanopterin, . This enzyme has been crystallized; it contains no prosthetic group A prosthetic group is the non-amino acid component that is part of the structure of the heteroproteins or conjugated proteins, being tightly l ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
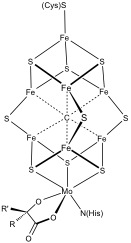
Iron–sulfur Protein
Iron–sulfur proteins are proteins characterized by the presence of iron–sulfur clusters containing sulfide-linked di-, tri-, and tetrairon centers in variable oxidation states. Iron–sulfur clusters are found in a variety of metalloproteins, such as the ferredoxins, as well as NADH dehydrogenase, hydrogenases, coenzyme Q – cytochrome c reductase, succinate – coenzyme Q reductase and nitrogenase. Iron–sulfur clusters are best known for their role in the oxidation-reduction reactions of electron transport in mitochondria and chloroplasts. Both Complex I and Complex II of oxidative phosphorylation have multiple Fe–S clusters. They have many other functions including catalysis as illustrated by aconitase, generation of radicals as illustrated by SAM-dependent enzymes, and as sulfur donors in the biosynthesis of lipoic acid and biotin. Additionally, some Fe–S proteins regulate gene expression. Fe–S proteins are vulnerable to attack by biogenic nitric oxide, formin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tetrahydrofolic Acid
Tetrahydrofolic acid (THFA), or tetrahydrofolate, is a folic acid derivative. Metabolism In humans, tetrahydrofolic acid is produced from dihydrofolic acid by dihydrofolate reductase. This reaction is inhibited by methotrexate. It is converted into 5,10-methylenetetrahydrofolate by serine hydroxymethyltransferase. Many bacteria produce tetrahydrofolic acid via dihydropteroate. Humans lack the enzymes to do this, thus molecules that shut down these enzymes are effective antibacterial compounds. For example, sulfonamide antibiotics competitively binds the active site of dihydropteroate synthetase, ecluding the binding of the dihydropteroate precuror, 4-aminobenzoic acid (PABA). Functions Tetrahydrofolic acid is a cofactor in many reactions, especially in the synthesis (or anabolism) of amino acids and nucleic acids. In addition, it serves as a carrier molecule for single-carbon moieties, that is, groups containing one carbon atom e.g. methyl, methylene, methenyl, formyl, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coenzyme M Methyltransferase
A cofactor is a non-protein chemical compound or metallic ion that is required for an enzyme's role as a catalyst (a catalyst is a substance that increases the rate of a chemical reaction). Cofactors can be considered "helper molecules" that assist in biochemical transformations. The rates at which these happen are characterized in an area of study called enzyme kinetics. Cofactors typically differ from ligands in that they often derive their function by remaining bound. Cofactors can be classified into two types: inorganic ions and complex organic molecules called coenzymes. Coenzymes are mostly derived from vitamins and other organic essential nutrients in small amounts. (Some scientists limit the use of the term "cofactor" for inorganic substances; both types are included here.) Coenzymes are further divided into two types. The first is called a "prosthetic group", which consists of a coenzyme that is tightly (or even covalently and, therefore, permanently) bound to a protei ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coenzyme
A cofactor is a non-protein chemical compound or Metal ions in aqueous solution, metallic ion that is required for an enzyme's role as a catalysis, catalyst (a catalyst is a substance that increases the rate of a chemical reaction). Cofactors can be considered "helper molecules" that assist in Biochemistry, biochemical transformations. The rates at which these happen are characterized in an area of study called enzyme kinetics. Cofactors typically differ from Ligand (biochemistry), ligands in that they often derive their function by remaining bound. Cofactors can be classified into two types: inorganic ions and complex organic molecules called Enzyme#Coenzymes, coenzymes. Coenzymes are mostly derived from vitamins and other organic essential nutrients in small amounts. (Some scientists limit the use of the term "cofactor" for inorganic substances; both types are included here.) Coenzymes are further divided into two types. The first is called a "prosthetic group", which consists ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methanogenesis
Methanogenesis or biomethanation is the formation of methane coupled to energy conservation by microbes known as methanogens. It is the fourth and final stage of anaerobic digestion. Organisms capable of producing methane for energy conservation have been identified only from the domain Archaea, a group phylogenetically distinct from both eukaryotes and bacteria, although many live in close association with anaerobic bacteria. The production of methane is an important and widespread form of microbial metabolism. In anoxic environments, it is the final step in the decomposition of biomass. Methanogenesis is responsible for significant amounts of natural gas accumulations, the remainder being thermogenic. Biochemistry Methanogenesis in microbes is a form of anaerobic respiration. Methanogens do not use oxygen to respire; in fact, oxygen inhibits the growth of methanogens. The terminal electron acceptor in methanogenesis is not oxygen, but carbon. The two best described pathways ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pterin
Pterin is a heterocyclic compound composed of a pteridine ring system, with a " keto group" (a lactam) and an amino group on positions 4 and 2 respectively. It is structurally related to the parent bicyclic heterocycle called pteridine. Pterins, as a group, are compounds related to pterin with additional substituents. Pterin itself is of no biological significance. Pterins were first discovered in the pigments of butterfly wings (hence the origin of their name, from the Greek ''pteron'' (), wing) and perform many roles in coloration in the biological world. Chemistry Pterins exhibit a wide range of tautomerism in water, beyond what is assumed by just keto-enol tautomerism. For the unsubstituted pterin, at least five tautomers are commonly cited. For 6-methylpterin, seven tautomers are theoretically predicted to be important in solution. The pteridine ring system contains four nitrogen atoms, reducing its aromaticity to the point that it can be attacked by nucleophi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |