|
TaqMan Probes
TaqMan probes are hydrolysis probes that are designed to increase the specificity of quantitative PCR. The method was first reported in 1991 by researcher Kary Mullis at Cetus Corporation, and the technology was subsequently developed by Hoffmann-La Roche for diagnostic assays and by Applied Biosystems (now part of Thermo Fisher Scientific) for research applications. The TaqMan probe principle relies on the 5´–3´ exonuclease activity of ''Taq'' polymerase to cleave a dual-labeled probe during hybridization to the complementary target sequence and fluorophore-based detection. As in other quantitative PCR methods, the resulting fluorescence signal permits quantitative measurements of the accumulation of the product during the exponential stages of the PCR; however, the TaqMan probe significantly increases the specificity of the detection. TaqMan probes were named after the videogame Pac-Man (''Taq'' Polymerase + PacMan = TaqMan) as its mechanism is based on the Pac-Man pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrolysis
Hydrolysis (; ) is any chemical reaction in which a molecule of water breaks one or more chemical bonds. The term is used broadly for substitution, elimination, and solvation reactions in which water is the nucleophile. Biological hydrolysis is the cleavage of biomolecules where a water molecule is consumed to effect the separation of a larger molecule into component parts. When a carbohydrate is broken into its component sugar molecules by hydrolysis (e.g., sucrose being broken down into glucose and fructose), this is recognized as saccharification. Hydrolysis reactions can be the reverse of a condensation reaction in which two molecules join into a larger one and eject a water molecule. Thus hydrolysis adds water to break down, whereas condensation builds up by removing water. Types Usually hydrolysis is a chemical process in which a molecule of water is added to a substance. Sometimes this addition causes both the substance and water molecule to split into two parts. I ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rhodamine
Rhodamine is a family of related dyes, a subset of the triarylmethane dyes. They are derivatives of xanthene. Important members of the rhodamine family are Rhodamine 6G, Rhodamine 123, and Rhodamine B. They are mainly used to dye paper and inks, but they lack the lightfastness for fabric dyeing. Use Aside from their major applications, they are often used as a tracer dye, e.g. to determine the rate and direction of flow and transport of water. Rhodamine dyes fluoresce and can thus be detected easily and inexpensively with instruments called fluorometers. Rhodamine dyes are used extensively in biotechnology applications such as fluorescence microscopy, flow cytometry, fluorescence correlation spectroscopy and ELISA. Rhodamine 123 is used in biochemistry to inhibit mitochondrion function. Rhodamine 123 appears to bind to the mitochondrial membranes and inhibit transport processes, especially the electron transport chain, thus slowing down cellular respiration. It is a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SYBR Green
SYBR Green I (SG) is an asymmetrical cyanine dye used as a nucleic acid stain in molecular biology. The SYBR family of dyes is produced by Molecular Probes Inc., now owned by Thermo Fisher Scientific. SYBR Green I binds to DNA. The resulting DNA-dye-complex best absorbs 497 nanometer blue light (λmax = 497 nm) and emits green light (λmax = 520 nm). The stain preferentially binds to double-stranded DNA, but will stain single-stranded (ss) DNA with lower performance. SYBR Green can also stain RNA with a lower performance than ssDNA. Uses SYBR Green finds usage in several areas of biochemistry and molecular biology. It is used as a dye for the quantification of double stranded DNA in some methods of quantitative PCR. It is also used to visualise DNA in gel electrophoresis. Higher concentrations of SYBR Green can be used to stain agarose gels in order to visualise the DNA present. In addition to labelling pure nucleic acids, SYBR Green can also be used for labelling ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
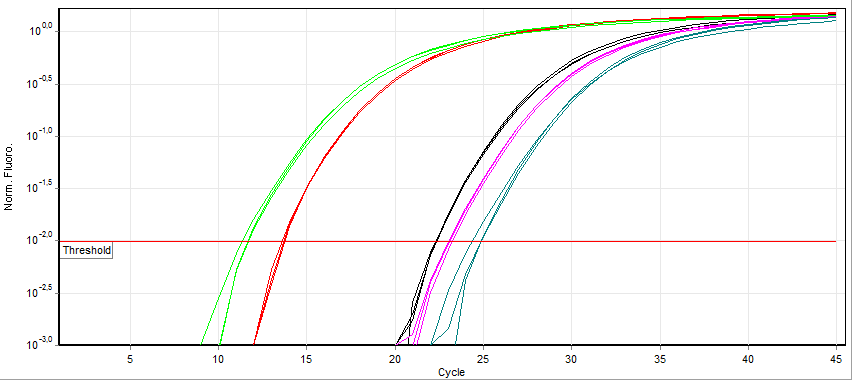
Quantitative PCR
A real-time polymerase chain reaction (real-time PCR, or qPCR) is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR (i.e., in real time), not at its end, as in conventional PCR. Real-time PCR can be used quantitatively (quantitative real-time PCR) and semi-quantitatively (i.e., above/below a certain amount of DNA molecules) (semi-quantitative real-time PCR). Two common methods for the detection of PCR products in real-time PCR are (1) non-specific fluorescent dyes that intercalate with any double-stranded DNA and (2) sequence-specific DNA probes consisting of oligonucleotides that are labelled with a fluorescent reporter, which permits detection only after hybridization of the probe with its complementary sequence. The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines propose that the abbreviation ''qPCR'' be used for q ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microarray
A microarray is a multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of genes from a sample (e.g. from a tissue). It is a two-dimensional array on a solid substrate—usually a glass slide or silicon thin-film cell—that assays (tests) large amounts of biological material using high-throughput screening miniaturized, multiplexed and parallel processing and detection methods. The concept and methodology of microarrays was first introduced and illustrated in antibody microarrays (also referred to as antibody matrix) by Tse Wen Chang in 1983 in a scientific publication and a series of patents. The " gene chip" industry started to grow significantly after the 1995 '' Science Magazine'' article by the Ron Davis and Pat Brown labs at Stanford University. With the establishment of companies, such as Affymetrix, Agilent, Applied Microarrays, Arrayjet, Illumina, and others, the technology of DNA microarrays has become the most sophis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SNP Genotyping
SNP genotyping is the measurement of genetic variations of single nucleotide polymorphisms (SNPs) between members of a species. It is a form of genotyping, which is the measurement of more general genetic variation. SNPs are one of the most common types of genetic variation. An SNP is a single base pair mutation at a specific locus, usually consisting of two alleles (where the rare allele frequency is > 1%). SNPs are found to be involved in the etiology of many human diseases and are becoming of particular interest in pharmacogenetics. Because SNPs are conserved during evolution, they have been proposed as markers for use in quantitative trait loci ( QTL) analysis and in association studies in place of microsatellites. The use of SNPs is being extended in the HapMap project, which aims to provide the minimal set of SNPs needed to genotype the human genome. SNPs can also provide a genetic fingerprint for use in identity testing. The increase of interest in SNPs has been reflect ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hepatitis
Hepatitis is inflammation of the liver parenchyma, liver tissue. Some people or animals with hepatitis have no symptoms, whereas others develop yellow discoloration of the skin and whites of the eyes (jaundice), Anorexia (symptom), poor appetite, vomiting, fatigue (medicine), tiredness, abdominal pain, and diarrhea. Hepatitis is ''acute (medicine), acute'' if it resolves within six months, and ''chronic condition, chronic'' if it lasts longer than six months. Acute hepatitis can self-limiting (biology), resolve on its own, progress to chronic hepatitis, or (rarely) result in acute liver failure. Chronic hepatitis may progress to scarring of the liver (cirrhosis), liver failure, and liver cancer. Hepatitis is most commonly caused by the virus ''hepatovirus A'', ''hepatitis B virus, B'', ''hepatitis C virus, C'', ''hepatitis D virus, D'', and ''hepatitis E virus, E''. Other Viral hepatitis, viruses can also cause liver inflammation, including cytomegalovirus, Epstein–Barr virus, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Human Leukocyte Antigen
The human leukocyte antigen (HLA) system or complex is a complex of genes on chromosome 6 in humans which encode cell-surface proteins responsible for the regulation of the immune system. The HLA system is also known as the human version of the major histocompatibility complex (MHC) found in many animals. Mutations in HLA genes may be linked to autoimmune disease such as type I diabetes, and celiac disease. The HLA gene complex resides on a 3 Mbp stretch within chromosome 6, p-arm at 21.3. HLA genes are highly polymorphic, which means that they have many different alleles, allowing them to fine-tune the adaptive immune system. The proteins encoded by certain genes are also known as ''antigens'', as a result of their historic discovery as factors in organ transplants. HLAs corresponding to MHC class I ( A, B, and C), all of which are the HLA Class1 group, present peptides from inside the cell. For example, if the cell is infected by a virus, the HLA system brings f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pharmacogenomics
Pharmacogenomics is the study of the role of the genome in drug response. Its name ('' pharmaco-'' + ''genomics'') reflects its combining of pharmacology and genomics. Pharmacogenomics analyzes how the genetic makeup of an individual affects their response to drugs. It deals with the influence of acquired and inherited genetic variation on drug response in patients by correlating DNA mutations (including single-nucleotide polymorphisms, copy number variations, and insertions/deletions) with pharmacokinetic (drug absorption, distribution, metabolism, and elimination), pharmacodynamic (effects mediated through a drug's biological targets), and/or immunogenic endpoints. Pharmacogenomics aims to develop rational means to optimize drug therapy, with respect to the patients' genotype, to ensure maximum efficiency with minimal adverse effects. Through the utilization of pharmacogenomics, it is hoped that pharmaceutical drug treatments can deviate from what is dubbed as the "one- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. Gene expression is summarized in the central dogma of molecular biology first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication. The process of gene expression is used by all known life— eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and utilized by viruses—to generate the macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Medical Laboratories
A medical laboratory or clinical laboratory is a laboratory where tests are conducted out on clinical specimens to obtain information about the health of a patient to aid in diagnosis, treatment, and prevention of disease. Clinical Medical laboratories are an example of applied science, as opposed to research laboratories that focus on basic science, such as found in some academic institutions. Medical laboratories vary in size and complexity and so offer a variety of testing services. More comprehensive services can be found in acute-care hospitals and medical centers, where 70% of clinical decisions are based on laboratory testing. Doctors offices and clinics, as well as skilled nursing and long-term care facilities, may have laboratories that provide more basic testing services. Commercial medical laboratories operate as independent businesses and provide testing that is otherwise not provided in other settings due to low test volume or complexity. Departments In ho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Thermal Cycler
The thermal cycler (also known as a thermocycler, PCR machine or DNA amplifier) is a laboratory apparatus most commonly used to amplify segments of DNA via the polymerase chain reaction (PCR). Thermal cyclers may also be used in laboratories to facilitate other temperature-sensitive reactions, including restriction enzyme digestion or rapid diagnostics. The device has a ''thermal block'' with holes where tubes holding the reaction mixtures can be inserted. The cycler then raises and lowers the temperature of the block in discrete, pre-programmed steps. History The earliest thermal cyclers were designed for use with the Klenow fragment of DNA polymerase I. Since this enzyme is destroyed during each heating step of the amplification process, new enzyme had to be added every cycle. This led to a cumbersome machine based on an automated pipettor, with open reaction tubes. Later, the PCR process was adapted to the use of thermostable DNA polymerase from '' Thermus aquaticus' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |