|
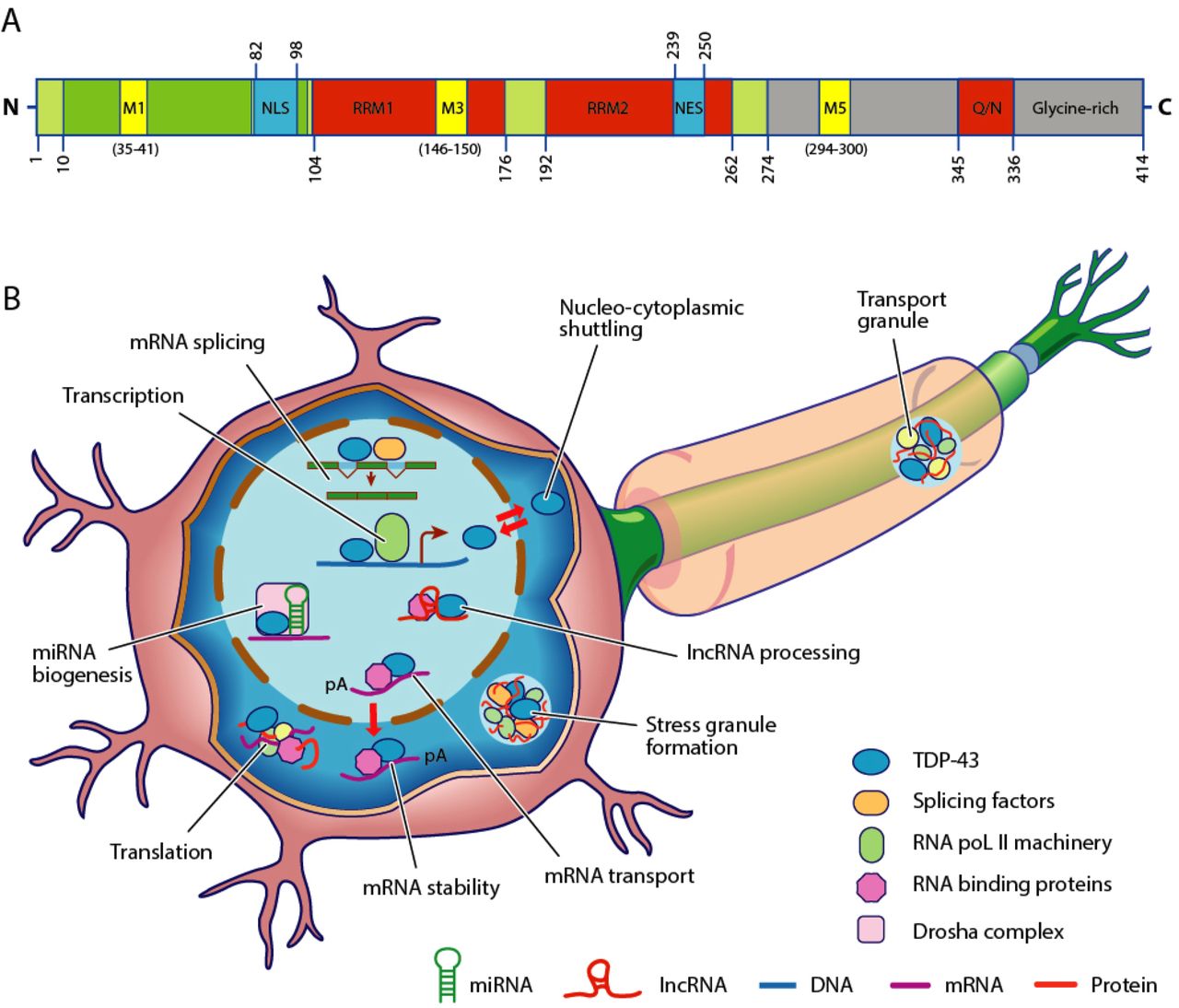
TDP-43
Transactive response DNA binding protein 43 kDa (TAR DNA-binding protein 43 or TDP-43) is a protein that in humans is encoded by the ''TARDBP'' gene. Structure TDP-43 is 414 amino acid residues long. It consists of four domains: an N-terminal domain spanning residues 1–76 (NTD) with a well-defined fold that has been shown to form a dimer or oligomer; two highly conserved folded RNA recognition motifs spanning residues 106–176 (RRM1) and 191–259 (RRM2), respectively, required to bind target RNA and DNA; an unstructured C-terminal domain encompassing residues 274–414 (CTD), which contains a glycine-rich region, is involved in protein-protein interactions, and harbors most of the mutations associated with familial amyotrophic lateral sclerosis. The entire protein devoid of large solubilising tags has been purified. The full-length protein is a dimer. The dimer is formed due to a self-interaction between two NTD domains, where the dimerisation can be propagated to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Frontotemporal Lobar Degeneration
Frontotemporal lobar degeneration (FTLD) is a pathological process that occurs in frontotemporal dementia. It is characterized by atrophy in the frontal lobe and temporal lobe of the brain, with sparing of the parietal and occipital lobes. Common proteinopathies that are found in FTLD include the accumulation of tau proteins and TAR DNA-binding protein 43 (TDP-43). Mutations in the '' C9orf72'' gene have been established as a major genetic contribution of FTLD, although defects in the granulin (GRN) and microtubule-associated proteins (MAPs) are also associated with it. Classification There are 3 main histological subtypes found at post-mortem: * FTLD-tau is characterised by tau positive inclusion bodies often referred to as Pick-bodies. Examples of FTLD-tau include; Pick's disease, corticobasal degeneration, progressive supranuclear palsy. * FTLD-TDP (or FTLD-U ) is characterised by ubiquitin and TDP-43 positive, tau negative, FUS negative inclusion bodies. The pat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TMEM106B
Transmembrane protein 106B is a protein that is encoded by the ''TMEM106B'' gene. It is found primarily within neurons and oligodendrocytes in the central nervous system with its subcellular location being in lysosomal membranes. TMEM106B helps facilitate important functions for maintaining a healthy lysosome, and therefore certain mutations and polymorphisms can lead to issues with proper lysosomal function. Lysosomes are in charge of clearing out mis-folded proteins and other debris, and thus, play an important role in neurodegenerative diseases that are driven by the accumulation of various mis-folded proteins and aggregates. Due to its impact on lysosomal function, TMEM106B has been investigated and found to be associated to multiple neurodegenerative diseases. Structure Gene In humans,''TMEM106B'' is found on chromosome 7 at positions 12211270 – 12243367, totaling 32097 base pairs. The gene includes 9 exons and can give rise to two different isoforms, T185 and S185, w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
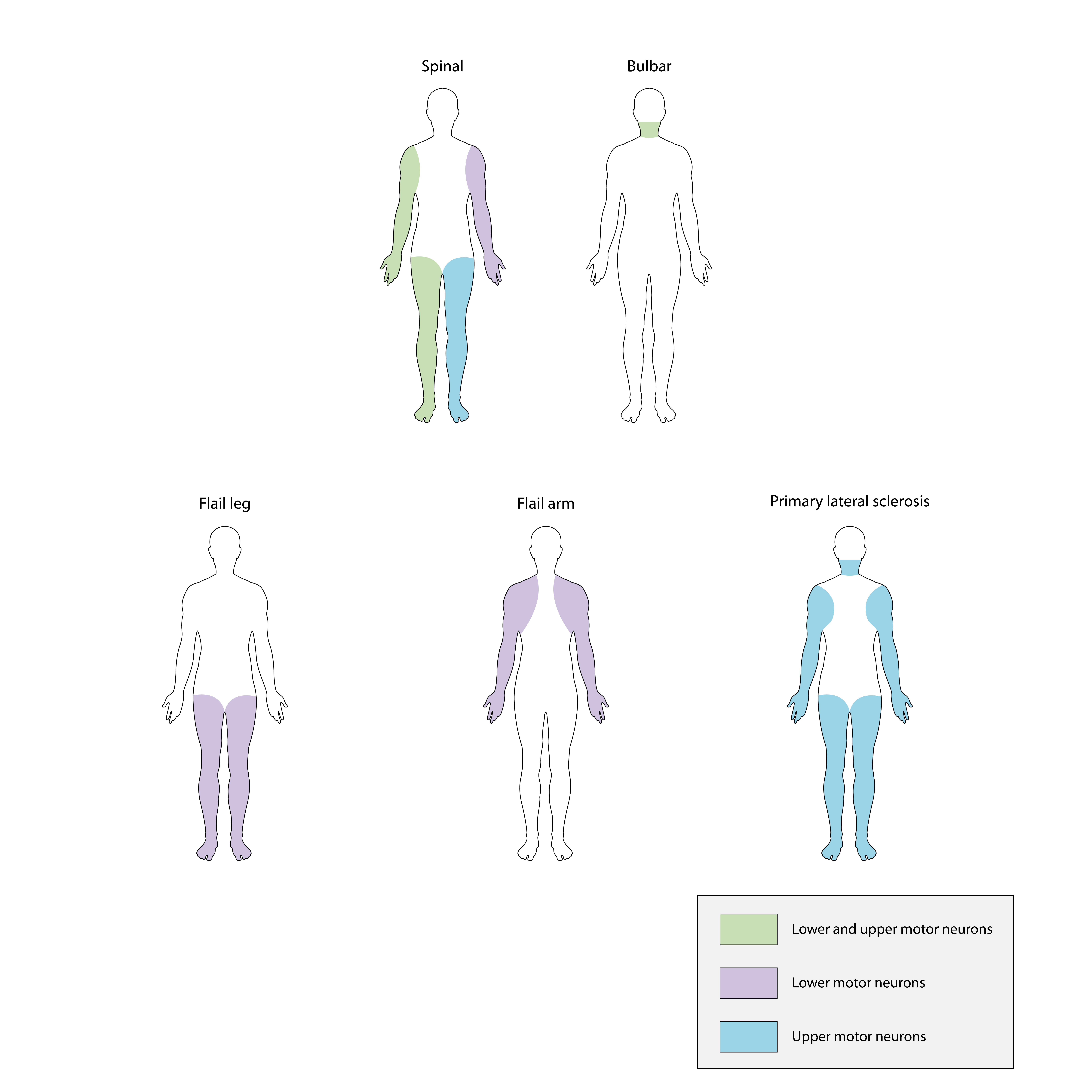
Amyotrophic Lateral Sclerosis
Amyotrophic lateral sclerosis (ALS), also known as motor neuron disease (MND) or—in the United States—Lou Gehrig's disease (LGD), is a rare, Terminal illness, terminal neurodegenerative disease, neurodegenerative disorder that results in the progressive loss of both upper and lower motor neurons that normally control Skeletal muscle, voluntary muscle contraction. ALS is the most common form of the motor neuron diseases. ALS often presents in its early stages with gradual muscle Spasticity, stiffness, Fasciculation, twitches, Muscle weakness, weakness, and Muscle atrophy, wasting. Motor neuron loss typically continues until the abilities to eat, speak, move, and, lastly, breathe are all lost. While only 15% of people with ALS also fully develop frontotemporal dementia, an estimated 50% face at least some minor difficulties with cognitive disorder, thinking and behavior. Depending on which of the aforementioned symptoms develops first, ALS is classified as ''limb-onset'' (b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Solenoid Protein Domain
Solenoid protein domains are a highly modular type of protein domain. They consist of a chain of nearly identical Protein structure, folds, often simply called Protein tandem repeats, tandem repeats. They are extremely common among all types of proteins, though exact figures are unknown. "Repeats" in molecular biology In proteins, a "repeat" is any sequence block that returns more than one time in the protein sequence, sequence, either in an identical or a highly similar form. Repetitiveness does not in itself indicate anything about the structure of the protein. As a "rule of thumb", short repetitive sequences (e.g. those below the length of 10 amino acids) may be intrinsically unstructured proteins, intrinsically disordered, and not part of any protein folding, folded protein domains. Repeats that are at least 30 to 40 amino acids long, are far more likely to be folded as part of a domain. Such long repeats are frequently indicative of the presence of a solenoid domain in the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ssdna
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of all known organisms and many viruses. DNA and ribonucleic acid (RNA) are nucleic acids. Alongside proteins, lipids and complex carbohydrates (polysaccharides), nucleic acids are one of the four major types of macromolecules that are essential for all known forms of life. The two DNA strands are known as polynucleotides as they are composed of simpler monomeric units called nucleotides. Each nucleotide is composed of one of four nitrogen-containing nucleobases (cytosine guanine adenine or thymine , a sugar called deoxyribose, and a phosphate group. The nucleotides are joined to one another in a chain by covalent bonds (known as the phosphodiester linkage) between the sugar of one nucleotide and the phosphate of the next, resulting in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sulfate
The sulfate or sulphate ion is a polyatomic anion with the empirical formula . Salts, acid derivatives, and peroxides of sulfate are widely used in industry. Sulfates occur widely in everyday life. Sulfates are salts of sulfuric acid and many are prepared from that acid. Spelling "Sulfate" is the spelling recommended by IUPAC, but "sulphate" was traditionally used in British English. Structure The sulfate anion consists of a central sulfur atom surrounded by four equivalent oxygen atoms in a tetrahedral arrangement. The symmetry of the isolated anion is the same as that of methane. The sulfur atom is in the +6 oxidation state while the four oxygen atoms are each in the −2 state. The sulfate ion carries an overall charge of −2 and it is the conjugate base of the bisulfate (or hydrogensulfate) ion, , which is in turn the conjugate base of , sulfuric acid. Organic sulfate esters, such as dimethyl sulfate, are covalent compounds and esters of sulfuric acid. The tetrahedral ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
An alpha helix (or α-helix) is a sequence of amino acids in a protein that are twisted into a coil (a helix). The alpha helix is the most common structural arrangement in the Protein secondary structure, secondary structure of proteins. It is also the most extreme type of local structure, and it is the local structure that is most easily predicted from a sequence of amino acids. The alpha helix has a right-handed helix conformation in which every backbone amino, N−H group hydrogen bonds to the backbone carbonyl, C=O group of the amino acid that is four residue (biochemistry), residues earlier in the protein sequence. Other names The alpha helix is also commonly called a: * Pauling–Corey–Branson α-helix (from the names of three scientists who described its structure) * 3.613-helix because there are 3.6 amino acids in one ring, with 13 atoms being involved in the ring formed by the hydrogen bond (starting with amidic hydrogen and ending with carbonyl oxygen) Discovery ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Sheet
The beta sheet (β-sheet, also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, Alzheimer's disease and other proteinopathies. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin Ligase
A ubiquitin ligase (also called an E3 ubiquitin ligase) is a protein that recruits an E2 ubiquitin-conjugating enzyme that has been loaded with ubiquitin, recognizes a protein substrate, and assists or directly catalyzes the transfer of ubiquitin from the E2 to the protein substrate. In simple and more general terms, the ligase enables movement of ubiquitin from a ubiquitin carrier to another protein (the substrate) by some mechanism. The ubiquitin, once it reaches its destination, ends up being attached by an isopeptide bond to a lysine residue, which is part of the target protein. E3 ligases interact with both the target protein and the E2 enzyme, and so impart substrate specificity to the E2. Commonly, E3s polyubiquitinate their substrate with Lys48-linked chains of ubiquitin, targeting the substrate for destruction by the proteasome. However, many other types of linkages are possible and alter a protein's activity, interactions, or localization. Ubiquitination by E3 ligases re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin-like Protein
Ubiquitin-like proteins (UBLs) are a family of small proteins involved in post-translational modification of other proteins in a cell (biology), cell, usually with a regulatory protein, regulatory function. The UBL protein family derives its name from the first member of the class to be discovered, ubiquitin (Ub), best known for its role in regulating protein degradation through covalent modification of other proteins. Following the discovery of ubiquitin, many additional evolutionarily related members of the group were described, involving parallel regulatory processes and similar chemistry. UBLs are involved in a widely varying array of cellular functions including autophagy, protein trafficking, inflammation and immune responses, transcription (biology), transcription, DNA repair, RNA splicing, and cellular differentiation. Discovery Ubiquitin itself was first discovered in the 1970s and originally named "ubiquitous immunopoietic polypeptide". Subsequently, other proteins with ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell (biology)
The cell is the basic structural and functional unit of all life, forms of life. Every cell consists of cytoplasm enclosed within a Cell membrane, membrane; many cells contain organelles, each with a specific function. The term comes from the Latin word meaning 'small room'. Most cells are only visible under a light microscope, microscope. Cells Abiogenesis, emerged on Earth about 4 billion years ago. All cells are capable of Self-replication, replication, protein synthesis, and cell motility, motility. Cells are broadly categorized into two types: eukaryotic cells, which possess a Cell nucleus, nucleus, and prokaryotic, prokaryotic cells, which lack a nucleus but have a nucleoid region. Prokaryotes are single-celled organisms such as bacteria, whereas eukaryotes can be either single-celled, such as amoebae, or multicellular organism, multicellular, such as some algae, plants, animals, and fungi. Eukaryotic cells contain organelles including Mitochondrion, mitochondria, which ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligomer
In chemistry and biochemistry, an oligomer () is a molecule that consists of a few repeating units which could be derived, actually or conceptually, from smaller molecules, monomers.Quote: ''Oligomer molecule: A molecule of intermediate relative molecular mass, the structure of which essentially comprises a small plurality of units derived, actually or conceptually, from molecules of lower relative molecular mass.'' The name is composed of Greek elements '' oligo-'', "a few" and '' -mer'', "parts". An adjective form is ''oligomeric''. The oligomer concept is contrasted to that of a polymer, which is usually understood to have a large number of units, possibly thousands or millions. However, there is no sharp distinction between these two concepts. One proposed criterion is whether the molecule's properties vary significantly with the removal of one or a few of the units. An oligomer with a specific number of units is referred to by the Greek prefix denoting that number, w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |