|
SSC Buffer
In biochemistry and molecular biology, saline-sodium citrate (SSC) buffer is used as a hybridization buffer, to control stringency for washing steps in protocols for Southern blotting, in situ hybridization, DNA Microarray or Northern blotting. 20X SSC may be used to prevent drying of agarose gels during a vacuum transfer. A 20X stock solution consists of 3 M sodium chloride and 300 mM trisodium citrate (adjusted to pH 7.0 with HCl). See also *Southern Blot *Northern Blot *DNA Microarray *DNA Fingerprinting DNA profiling (also called DNA fingerprinting) is the process of determining an individual's DNA characteristics. DNA analysis intended to identify a species, rather than an individual, is called DNA barcoding. DNA profiling is a forensic te ... References Material safety data sheet for 20x SSC buffer, Mediatech, Inc. Buffer solutions {{biochem-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biochemistry
Biochemistry or biological chemistry is the study of chemical processes within and relating to living organisms. A sub-discipline of both chemistry and biology, biochemistry may be divided into three fields: structural biology, enzymology and metabolism. Over the last decades of the 20th century, biochemistry has become successful at explaining living processes through these three disciplines. Almost all areas of the life sciences are being uncovered and developed through biochemical methodology and research.Voet (2005), p. 3. Biochemistry focuses on understanding the chemical basis which allows biological molecules to give rise to the processes that occur within living cells and between cells, Karp (2009), p. 2. in turn relating greatly to the understanding of tissues and organs, as well as organism structure and function. Miller (2012). p. 62. Biochemistry is closely related to molecular biology, which is the study of the molecular mechanisms of biological phenom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physical structure of biological macromolecules is known as molecular biology. Molecular biology was first described as an approach focused on the underpinnings of biological phenomena - uncovering the structures of biological molecules as well as their interactions, and how these interactions explain observations of classical biology. In 1945 the term molecular biology was used by physicist William Astbury. In 1953 Francis Crick, James Watson, Rosalind Franklin, and colleagues, working at Medical Research Council unit, Cavendish laboratory, Cambridge (now the MRC Laboratory of Molecular Biology), made a double helix model of DNA which changed the entire research scenario. They proposed the DNA structure based on previous research done by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Buffer (chemistry)
A buffer solution (more precisely, pH buffer or hydrogen ion buffer) is an aqueous solution consisting of a mixture of a weak acid and its conjugate base, or vice versa. Its pH changes very little when a small amount of strong acid or base is added to it. Buffer solutions are used as a means of keeping pH at a nearly constant value in a wide variety of chemical applications. In nature, there are many living systems that use buffering for pH regulation. For example, the bicarbonate buffering system is used to regulate the pH of blood, and bicarbonate also acts as a buffer in the ocean. Principles of buffering Buffer solutions resist pH change because of a chemical equilibrium between the weak acid HA and its conjugate base A−: When some strong acid is added to an equilibrium mixture of the weak acid and its conjugate base, hydrogen ions (H+) are added, and the equilibrium is shifted to the left, in accordance with Le Chatelier's principle. Because of this, the hydrogen i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Southern Blotting
A Southern blot is a method used in molecular biology for detection of a specific DNA sequence in DNA samples. Southern blotting combines transfer of electrophoresis-separated DNA fragments to a filter membrane and subsequent fragment detection by probe hybridization. The method is named after the British biologist Edwin Southern, who first published it in 1975. Other blotting methods (i.e., western blot, northern blot, eastern blot, southwestern blot) that employ similar principles, but using RNA or protein, have later been named in reference to Edwin Southern's name. As the label is eponymous, Southern is capitalised, as is conventional of proper nouns. The names for other blotting methods may follow this convention, by analogy. Method #Restriction endonucleases are used to cut high-molecular-weight DNA strands into smaller fragments. #The DNA fragments are then electrophoresed on an agarose gel to separate them by size. # If some of the DNA fragments are larger than ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
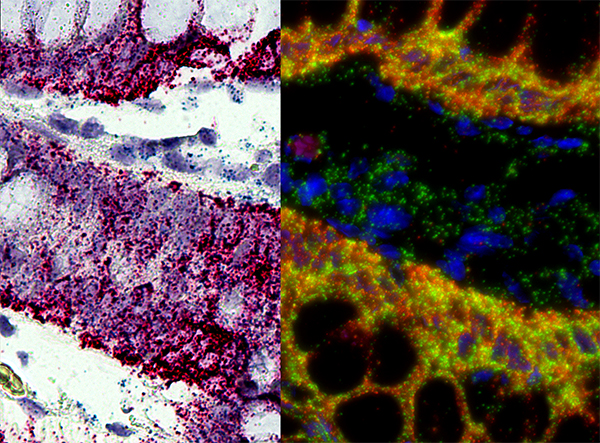
In Situ Hybridization
''In situ'' hybridization (ISH) is a type of hybridization that uses a labeled complementary DNA, RNA or modified nucleic acids strand (i.e., probe) to localize a specific DNA or RNA sequence in a portion or section of tissue (''in situ'') or if the tissue is small enough (e.g., plant seeds, ''Drosophila'' embryos), in the entire tissue (whole mount ISH), in cells, and in circulating tumor cells (CTCs). This is distinct from immunohistochemistry, which usually localizes proteins in tissue sections. In situ hybridization is used to reveal the location of specific nucleic acid sequences on chromosomes or in tissues, a crucial step for understanding the organization, regulation, and function of genes. The key techniques currently in use include ''in situ'' hybridization to mRNA with oligonucleotide and RNA probes (both radio-labeled and hapten-labeled), analysis with light and electron microscopes, whole mount ''in situ'' hybridization, double detection of RNAs and RNA plus prot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Microarray
A DNA microarray (also commonly known as DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles (10−12 moles) of a specific DNA sequence, known as '' probes'' (or ''reporters'' or '' oligos''). These can be a short section of a gene or other DNA element that are used to hybridize a cDNA or cRNA (also called anti-sense RNA) sample (called ''target'') under high-stringency conditions. Probe-target hybridization is usually detected and quantified by detection of fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. The original nucleic acid arrays were macro arrays approximately 9 cm × 12 cm and the first computerized image based analysis was published in 1981. It was i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
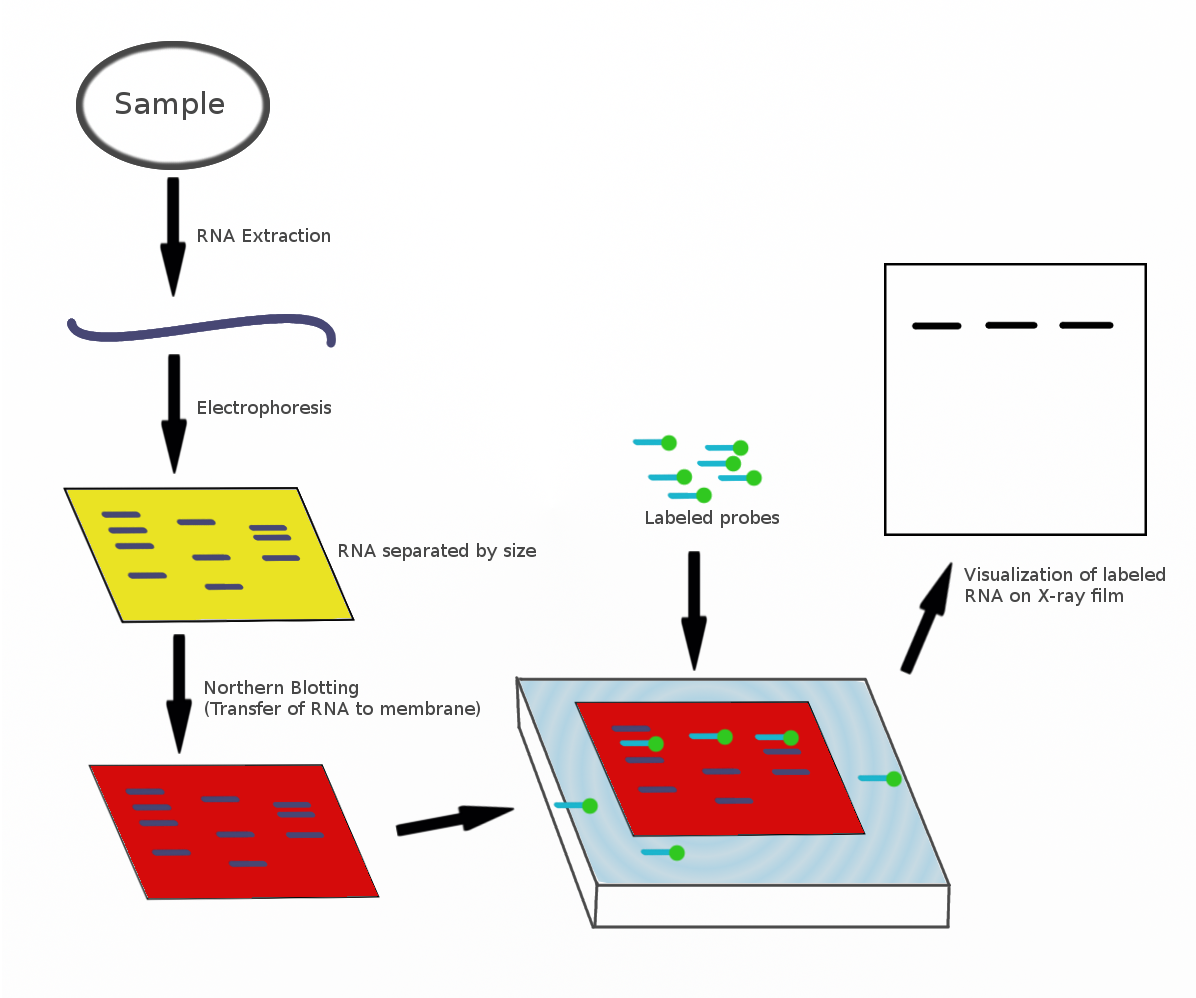
Northern Blotting
The northern blot, or RNA blot,Gilbert, S. F. (2000) Developmental Biology, 6th Ed. Sunderland MA, Sinauer Associates. is a technique used in molecular biology research to study gene expression by detection of RNA (or isolated mRNA) in a sample.Kevil, C. G., Walsh, L., Laroux, F. S., Kalogeris, T., Grisham, M. B., Alexander, J. S. (1997) An Improved, Rapid Northern Protocol. Biochem. and Biophys. Research Comm. 238:277–279. With northern blotting it is possible to observe cellular control over structure and function by determining the particular gene expression rates during differentiation and morphogenesis, as well as in abnormal or diseased conditions. Northern blotting involves the use of electrophoresis to separate RNA samples by size, and detection with a hybridization probe complementary to part of or the entire target sequence. Strictly speaking, the term 'northern blot' refers specifically to the capillary transfer of RNA from the electrophoresis gel to the blotting ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Agarose Gel
Agarose gel electrophoresis is a method of gel electrophoresis used in biochemistry, molecular biology, genetics, and clinical chemistry to separate a mixed population of macromolecules such as DNA or proteins in a matrix of agarose, one of the two main components of agar. The proteins may be separated by charge and/or size (isoelectric focusing agarose electrophoresis is essentially size independent), and the DNA and RNA fragments by length. Biomolecules are separated by applying an electric field to move the charged molecules through an agarose matrix, and the biomolecules are separated by size in the agarose gel matrix. Agarose gel is easy to cast, has relatively fewer charged groups, and is particularly suitable for separating DNA of size range most often encountered in laboratories, which accounts for the popularity of its use. The separated DNA may be viewed with stain, most commonly under UV light, and the DNA fragments can be extracted from the gel with relative ease. Mo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molar (concentration)
Molar concentration (also called molarity, amount concentration or substance concentration) is a measure of the concentration of a chemical species, in particular of a solute in a solution, in terms of amount of substance per unit volume of solution. In chemistry, the most commonly used unit for molarity is the number of moles per liter, having the unit symbol mol/L or mol/ dm3 in SI unit. A solution with a concentration of 1 mol/L is said to be 1 molar, commonly designated as 1 M. Definition Molar concentration or molarity is most commonly expressed in units of moles of solute per litre of solution. For use in broader applications, it is defined as amount of substance of solute per unit volume of solution, or per unit volume available to the species, represented by lowercase c: :c = \frac = \frac = \frac. Here, n is the amount of the solute in moles, N is the number of constituent particles present in volume V (in litres) of the solution, and N_\text is the Av ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sodium Chloride
Sodium chloride , commonly known as salt (although sea salt also contains other chemical salts), is an ionic compound with the chemical formula NaCl, representing a 1:1 ratio of sodium and chloride ions. With molar masses of 22.99 and 35.45 g/mol respectively, 100 g of NaCl contains 39.34 g Na and 60.66 g Cl. Sodium chloride is the salt most responsible for the salinity of seawater and of the extracellular fluid of many multicellular organisms. In its edible form, salt (also known as '' table salt'') is commonly used as a condiment and food preservative. Large quantities of sodium chloride are used in many industrial processes, and it is a major source of sodium and chlorine compounds used as feedstocks for further chemical syntheses. Another major application of sodium chloride is de-icing of roadways in sub-freezing weather. Uses In addition to the familiar domestic uses of salt, more dominant applications of the approximately 250 million tonnes per year productio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Milli-
''Milli'' (symbol m) is a unit prefix in the metric system denoting a factor of one thousandth (10−3). Proposed in 1793, and adopted in 1795, the prefix comes from the Latin , meaning ''one thousand'' (the Latin plural is ). Since 1960, the prefix is part of the International System of Units (SI). See also * RKM code The RKM code, also referred to as "letter and numeral code for resistance and capacitance values and tolerances", "letter and digit code for resistance and capacitance values and tolerances", or informally as "R notation" is a notation to specify ... References {{reflist SI prefixes 1000 (number) simple:Milli- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Trisodium Citrate
Trisodium citrate has the chemical formula of Na3C6H5O7. It is sometimes referred to simply as "sodium citrate", though sodium citrate can refer to any of the three sodium salts of citric acid. It possesses a saline, mildly tart flavor, and is a mild alkali. Applications Foods Sodium citrate is chiefly used as a food additive, usually for flavor or as a preservative. Its E number is E331. Sodium citrate is employed as a flavoring agent in certain varieties of club soda. It is common as an ingredient in bratwurst, and is also used in commercial ready-to-drink beverages and drink mixes, contributing a tart flavor. It is found in gelatin mix, ice cream, yogurt, jams, sweets, milk powder, processed cheeses, carbonated beverages, and wine, amongst others. Sodium citrate can be used as an emulsifying stabilizer when making cheese. It allows the cheese to melt without becoming greasy by stopping the fats from separating. Buffering As a conjugate base of a weak acid, citrate can p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |


.jpg)