|
SCOPE (protein Engineering)
Structure-based combinatorial protein engineering (SCOPE) is a synthetic biology technique for creating gene libraries ( lineages) of defined composition designed from structural and probabilistic constraints of the encoded proteins. The development of this technique was driven by fundamental questions about protein structure, function, and evolution, although the technique is generally applicable for the creation of engineered proteins with commercially desirable properties. Combinatorial travel through sequence spacetime is the goal of SCOPE. Description At its inception, SCOPE was developed as a homology-independent recombination technique to enable the creation of multiple crossover libraries from distantly related genes. In this application, an “exon plate tectonics” design strategy was devised to assemble “equivalent” elements of structure (continental plates) with variability in the junctions linking them ( fault lines) to explore global protein space. To create the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Synthetic Biology
Synthetic biology (SynBio) is a multidisciplinary field of science that focuses on living systems and organisms. It applies engineering principles to develop new biological parts, devices, and systems or to redesign existing systems found in nature. It is a branch of science that encompasses a broad range of methodologies from various disciplines, such as biochemistry, biotechnology, biomaterials, Materials science, material science/engineering, genetic engineering, molecular biology, molecular engineering, systems biology, Model lipid bilayer, membrane science, biophysics, Biological engineering, chemical and biological engineering, Electrical engineering, electrical and computer engineering, control engineering and evolutionary biology. It includes designing and constructing BioBrick, biological modules, biological systems, and biological machines, or re-designing existing biological systems for useful purposes. Additionally, it is the branch of science that focuses on the ne ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology (physical form and structure), its developmental processes, its biochemical and physiological properties, and its behavior. An organism's phenotype results from two basic factors: the expression of an organism's genetic code (its genotype) and the influence of environmental factors. Both factors may interact, further affecting the phenotype. When two or more clearly different phenotypes exist in the same population of a species, the species is called polymorphic. A well-documented example of polymorphism is Labrador Retriever coloring; while the coat color depends on many genes, it is clearly seen in the environment as yellow, black, and brown. Richard Dawkins in 1978 and again in his 1982 book '' The Extended Phenotype'' suggested that one can regard bird nests and other built structures such as caddisfly larva cases and beaver dams ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Design
Protein design is the rational design of new protein molecules to design novel activity, behavior, or purpose, and to advance basic understanding of protein function. Proteins can be designed from scratch (''de novo'' design) or by making calculated variants of a known protein structure and its sequence (termed ''protein redesign''). Rational protein design approaches make protein-sequence predictions that will fold to specific structures. These predicted sequences can then be validated experimentally through methods such as peptide synthesis, site-directed mutagenesis, or artificial gene synthesis. Rational protein design dates back to the mid-1970s. Recently, however, there were numerous examples of successful rational design of water-soluble and even transmembrane peptides and proteins, in part due to a better understanding of different factors contributing to protein structure stability and development of better computational methods. Overview and history The goal in ratio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid Analogues
Nucleic acid analogues are compounds which are analogous (structurally similar) to naturally occurring RNA and DNA, used in medicine and in molecular biology research. Nucleic acids are chains of nucleotides, which are composed of three parts: a phosphate backbone, a pentose sugar, either ribose or deoxyribose, and one of four nucleobases. An analogue may have any of these altered. Typically the analogue nucleobases confer, among other things, different base pairing and base stacking properties. Examples include universal bases, which can pair with all four canonical bases, and phosphate-sugar backbone analogues such as PNA, which affect the properties of the chain (PNA can even form a triple helix). Nucleic acid analogues are also called xeno nucleic acids and represent one of the main pillars of xenobiology, the design of new-to-nature forms of life based on alternative biochemistries. Artificial nucleic acids include peptide nucleic acids (PNA), morpholino, and locked nu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences (see non-coding DNA), and often a substantial fraction of junk DNA with no evident function. Almost all eukaryotes have mitochondrial DNA, mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplast DNA, chloroplasts with a chloroplast genome. The study of the genome is called genomics. The genomes of many organisms have been Whole-genome sequencing, sequenced and various regions have been annotated. The first genome to be sequenced was that of the virus φX174 in 1977; the first genome sequence of a prokaryote (''Haemophilus influenzae'') was published in 1995; the yeast (''Saccharomyces cerevisiae'') genome was the first eukaryotic genome to be sequenced in 1996. The Human Genome Project ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Synthesis
Artificial gene synthesis, or simply gene synthesis, refers to a group of methods that are used in synthetic biology to construct and assemble genes from nucleotides '' de novo''. Unlike DNA synthesis in living cells, artificial gene synthesis does not require template DNA, allowing virtually any DNA sequence to be synthesized in the laboratory. It comprises two main steps, the first of which is solid-phase DNA synthesis, sometimes known as DNA printing. This produces oligonucleotide fragments that are generally under 200 base pairs. The second step then involves connecting these oligonucleotide fragments using various DNA assembly methods. Because artificial gene synthesis does not require template DNA, it is theoretically possible to make a completely synthetic DNA molecule with no limits on the nucleotide sequence or size. Synthesis of the first complete gene, a yeast tRNA, was demonstrated by Har Gobind Khorana and coworkers in 1972. Synthesis of the first peptide- and protein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Expanded Genetic Code
An expanded genetic code is an artificially modified genetic code in which one or more specific codons have been re-allocated to encode an amino acid that is not among the 22 common naturally-encoded proteinogenic amino acids. The key prerequisites to expand the genetic code are: * the non-standard amino acid to encode, * an unused codon to adopt, * a tRNA that recognizes this codon, and * a tRNA synthetase that recognizes only that tRNA and only the non-standard amino acid. Expanding the genetic code is an area of research of synthetic biology, an applied biological discipline whose goal is to engineer living systems for useful purposes. The genetic code expansion enriches the repertoire of useful tools available to science. In May 2019, researchers, in a milestone effort, reported the creation of a new synthetic (possibly artificial) form of viable life, a variant of the bacteria ''Escherichia coli'', by reducing the natural number of 64 codons in the bacterial genome to 61 c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzymology
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts include Ribozyme, catalytic RNA molecules, also called ribozymes. They are sometimes descr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
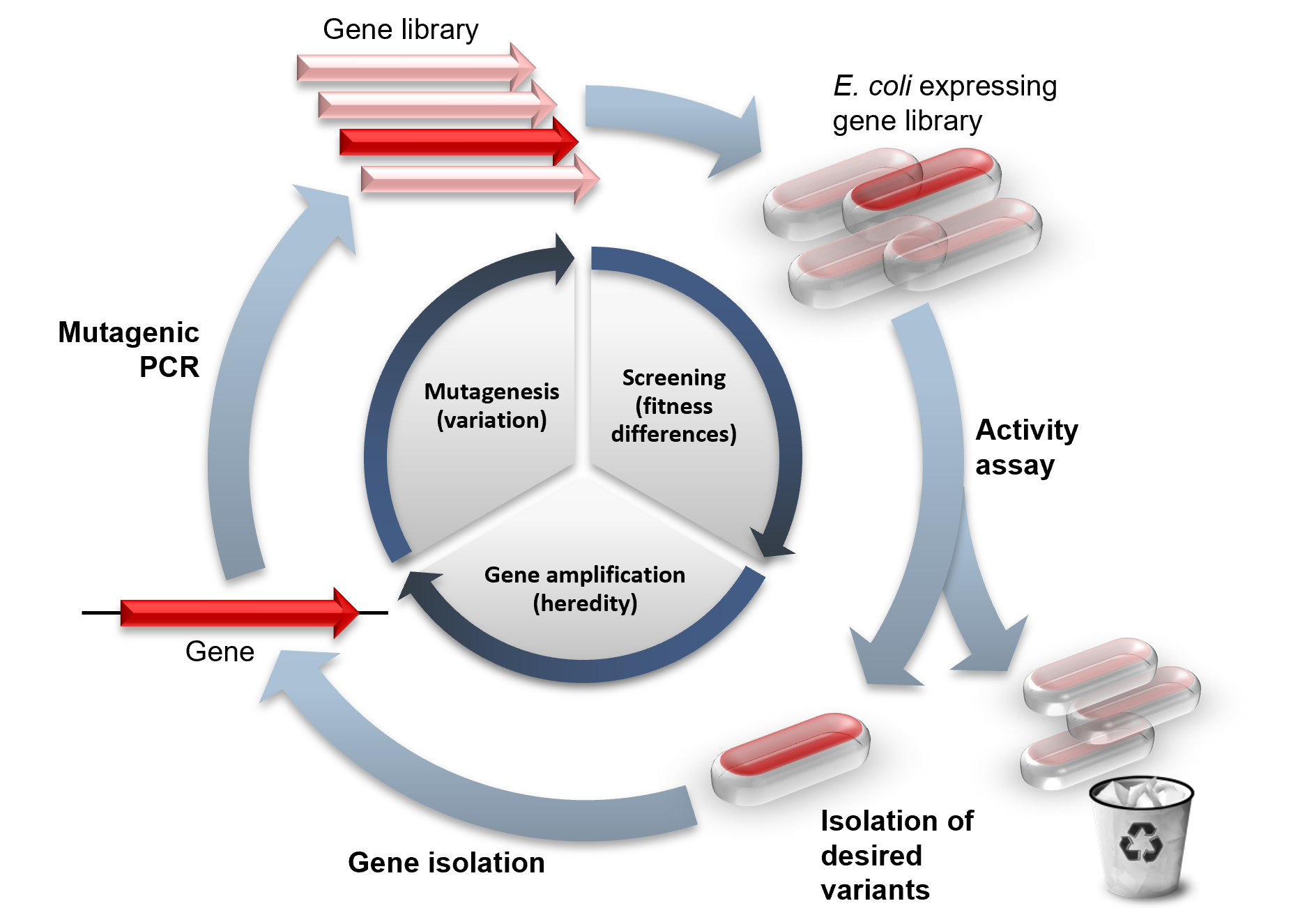
Directed Evolution
Directed evolution (DE) is a method used in protein engineering that mimics the process of natural selection to steer proteins or nucleic acids toward a user-defined goal. It consists of subjecting a gene to iterative rounds of mutagenesis (creating a library of variants), selection (expressing those variants and isolating members with the desired function) and amplification (generating a template for the next round). It can be performed ''in vivo'' (in living organisms), or ''in vitro'' (in cells or free in solution). Directed evolution is used both for protein engineering as an alternative to rationally designing modified proteins, as well as for experimental evolution studies of fundamental evolutionary principles in a controlled, laboratory environment. History Directed evolution has its origins in the 1960s with the evolution of RNA molecules in the "Spiegelman's Monster" experiment. The concept was extended to protein evolution via evolution of bacteria under selec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetics
In biology, phylogenetics () is the study of the evolutionary history of life using observable characteristics of organisms (or genes), which is known as phylogenetic inference. It infers the relationship among organisms based on empirical data and observed heritable traits of DNA sequences, protein amino acid sequences, and morphology. The results are a phylogenetic tree—a diagram depicting the hypothetical relationships among the organisms, reflecting their inferred evolutionary history. The tips of a phylogenetic tree represent the observed entities, which can be living taxa or fossils. A phylogenetic diagram can be rooted or unrooted. A rooted tree diagram indicates the hypothetical common ancestor of the taxa represented on the tree. An unrooted tree diagram (a network) makes no assumption about directionality of character state transformation, and does not show the origin or "root" of the taxa in question. In addition to their use for inferring phylogenetic pa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Saltation (biology)
In biology, saltation () is a sudden and large mutational change from one generation to the next, potentially causing single-step speciation. This was historically offered as an alternative to Darwinism. Some forms of mutationism were effectively saltationist, implying large discontinuous jumps. Speciation, such as by polyploidy in plants, can sometimes be achieved in a single and in evolutionary terms sudden step. Evidence exists for various forms of saltation in a variety of organisms. History Prior to Charles Darwin most evolutionary scientists had been saltationists. Jean-Baptiste Lamarck was a gradualist but similar to other scientists of the period had written that saltational evolution was possible. Étienne Geoffroy Saint-Hilaire endorsed a theory of saltational evolution that "monstrosities could become the founding fathers (or mothers) of new species by instantaneous transition from one form to the next." Geoffroy wrote that environmental pressures could produce sudde ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gradualism
Gradualism, from the Latin ("step"), is a hypothesis, a theory or a tenet assuming that change comes about gradually or that variation is gradual in nature and happens over time as opposed to in large steps. Uniformitarianism, incrementalism, and reformism are similar concepts. Gradualism can also refer to desired, controlled change in society, institutions, or policies. For example, social democrats and democratic socialists see the socialist society as achieved through gradualism. Geology and biology In the natural sciences, gradualism is the theory which holds that profound change is the cumulative product of slow but continuous processes, often contrasted with catastrophism. The theory was proposed in 1795 by James Hutton, a Scottish geologist, and was later incorporated into Charles Lyell's theory of uniformitarianism. Tenets from both theories were applied to biology and formed the basis of early evolutionary theory. Charles Darwin was influenced by Lyell's ''P ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |