|
RNA Polymerase III
In eukaryote cells, RNA polymerase III (also called Pol III) is a protein that transcribes DNA to synthesize ribosomal 5S rRNA, tRNA and other small RNAs. The genes transcribed by RNA Pol III fall in the category of "housekeeping" genes whose expression is required in all cell types and most environmental conditions. Therefore, the regulation of Pol III transcription is primarily tied to the regulation of cell growth and the cell cycle, thus requiring fewer regulatory proteins than RNA polymerase II. Under stress conditions however, the protein Maf1 represses Pol III activity. Rapamycin is another Pol III inhibitor via its direct target TOR. Transcription The process of transcription (by any polymerase) involves three main stages: *Initiation, requiring construction of the RNA polymerase complex on the gene's promoter *Elongation, the synthesis of the RNA transcript *Termination, the finishing of RNA transcription and disassembly of the RNA polymerase complex Initiation I ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription (genetics)
Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1–3% of total RNA samples. Less than 2% of the human genome can be transcribed into mRNA ( Human genome#Coding vs. noncoding DNA), while at least 80% of mammalian genomic DNA can be actively transcribed (in one or more types of cells), with the majority of this 80% considered to be ncRNA. Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript. Transcription proceeds in the following general steps: # RNA polymerase, together with one or more general transcription factors, binds to promoter ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
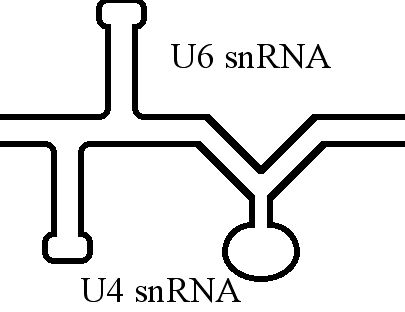
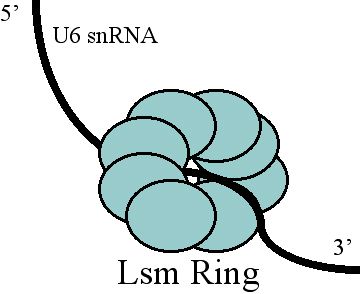
U6 SnRNA
U6 snRNA is the non-coding small nuclear RNA (snRNA) component of U6 snRNP (''small nuclear ribonucleoprotein''), an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex that catalyzes the excision of introns from pre-mRNA. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification and takes place only in the nucleus of eukaryotes. The RNA sequence of U6 is the most highly conserved across species of all five of the snRNAs involved in the spliceosome, suggesting that the function of the U6 snRNA has remained both crucial and unchanged through evolution. It is common in vertebrate genomes to find many copies of the U6 snRNA gene or U6-derived pseudogenes. This prevalence of "back-ups" of the U6 snRNA gene in vertebrates further implies its evolutionary importance to organism viability. The U6 snRNA gene has been isolated in many organi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
7SL RNA
The signal recognition particle RNA, (also known as 7SL, 6S or 4.5S RNA) is part of the signal recognition particle (SRP) ribonucleoprotein complex. SRP recognizes the signal peptide and binds to the ribosome, halting protein synthesis. Signal recognition particle receptor, is a protein that is embedded in a membrane, and which contains a Transmembrane channels, transmembrane pore. When the complex binds to , SRP releases the ribosome and drifts away. The ribosome resumes protein synthesis, but now the protein is moving through the transmembrane pore. In this way SRP directs the movement of proteins within the Cell (biology), cell to bind with a transmembrane pore which allows the protein to cross the membrane to where it is needed. The RNA and protein components of this complex are highly Conserved sequence, conserved but do vary between the different Kingdom (biology), kingdoms of life. The common Short interspersed nuclear element, SINE family Alu sequence, Alu probably or ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase MRP
RNase MRP (also called RMRP) is an enzymatically active ribonucleoprotein with two distinct roles in eukaryotes. RNAse MRP stands for RNAse for mitochondrial RNA processing. In mitochondria it plays a direct role in the initiation of mitochondrial DNA replication. In the nucleus it is involved in precursor rRNA processing, where it cleaves the internal transcribed spacer 1 between 18S and 5.8S rRNAs. Despite distinct functions, RNase MRP has been shown to be evolutionarily related to RNase P. Like eukaryotic RNase P, RNase MRP is not catalytically active without associated protein subunits. Mutations in the RNA component of RNase MRP cause cartilage–hair hypoplasia, a pleiotropic human disease. Responsible for this disease is a mutation in the RNase MRP RNA gene (RMRP), a non-coding RNA gene. RMRP was the first non-coding nuclear RNA gene found to cause disease. Mechanism and mutation effects RNase MRP and its role in pre-rRNA processing has been previously studied i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase P
Ribonuclease P (, ''RNase P'') is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein-based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA molecules. Further, RNase P is one of two known multiple turnover ribozymes in nature (the other being the ribosome), the discovery of which earned Sidney Altman and Thomas Cech the Nobel Prize in Chemistry in 1989: in the 1970s, Altman discovered the existence of precursor tRNA with flanking sequences and was the first to characterize RNase P and its activity in processing of the 5' leader sequence of precursor tRNA. Recent findings also reveal that RNase P has a new function. It has been shown that human nuclear RNase P is required for the normal and efficient transcription of various small noncoding RNAs, such as tRNA, 5S rRNA, SRP RNA and U6 snRNA genes, which ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
U6 Spliceosomal RNA
U6 snRNA is the non-coding small nuclear RNA (snRNA) component of U6 snRNP (''small nuclear ribonucleoprotein''), an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex that catalyzes the excision of introns from pre-mRNA. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification and takes place only in the nucleus of eukaryotes. The RNA sequence of U6 is the most highly conserved across species of all five of the snRNAs involved in the spliceosome, suggesting that the function of the U6 snRNA has remained both crucial and unchanged through evolution. It is common in vertebrate genomes to find many copies of the U6 snRNA gene or U6-derived pseudogenes. This prevalence of "back-ups" of the U6 snRNA gene in vertebrates further implies its evolutionary importance to organism viability. The U6 snRNA gene has been isolated in many org ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
5S Ribosomal RNA
The 5S ribosomal RNA (5S rRNA) is an approximately 120 nucleotide-long ribosomal RNA molecule with a mass of 40 kDa. It is a structural and functional component of the large subunit of the ribosome in all domains of life (bacteria, archaea, and eukaryotes), with the exception of mitochondrial ribosomes of fungi and animals. The designation 5S refers to the molecule's sedimentation velocity in an ultracentrifuge, which is measured in Svedberg units (S). Biosynthesis In prokaryotes, the 5S rRNA gene is typically located in the rRNA operons downstream of the small and the large subunit rRNA, and co-transcribed into a polycistronic precursor. A particularity of eukaryotic nuclear genomes is the occurrence of multiple 5S rRNA gene copies (5S rDNA) clustered in tandem repeats, with copy number varying from species to species. Eukaryotic 5S rRNA is synthesized by RNA polymerase III, whereas other eukaryotic rRNAs are cleaved from a 45S precursor transcribed by RNA polymerase I ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transfer RNA
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino acid sequence of proteins. tRNAs genes from Bacteria are typically shorter (mean = 77.6 bp) than tRNAs from Archaea (mean = 83.1 bp) and eukaryotes (mean = 84.7 bp). The mature tRNA follows an opposite pattern with tRNAs from Bacteria being usually longer (median = 77.6 nt) than tRNAs from Archaea (median = 76.8 nt), with eukaryotes exhibiting the shortest mature tRNAs (median = 74.5 nt). Transfer RNA (tRNA) does this by carrying an amino acid to the protein synthesizing machinery of a cell called the ribosome. Complementation of a 3-nucleotide codon in a messenger RNA (mRNA) by a 3-nucleotide anticodon of the tRNA results in protein synthesis based on the mRNA code. As such, tRNAs are a necessary component of translation, the biological s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The resulting structure is a key building block of many RNA secondary structures. As an important secondary structure of RNA, it can direct RNA folding, protect structural stability for messenger RNA (mRNA), provide recognition sites for RNA binding proteins, and serve as a substrate for enzymatic reactions. Formation and stability The formation of a stem-loop structure is dependent on the stability of the resulting helix and loop regions. The first prerequisite is the presence of a sequence that can fold back on itself to form a paired double helix. The stability of this helix is determined by its length, the number of mismatches or bulges it ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SNAPC5
snRNA-activating protein complex subunit 5 is a protein that in humans is encoded by the ''SNAPC5'' gene In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b .... References Further reading * * * * * * External links * * {{gene-15-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SNAPC4
snRNA-activating protein complex subunit 4 is a protein that in humans is encoded by the ''SNAPC4'' gene. Model organisms Model organisms have been used in the study of SNAPC4 function. A conditional knockout mouse line, called ''Snapc4tm1a(KOMP)Wtsi'' was generated as part of the International Knockout Mouse Consortium program — a high-throughput mutagenesis project to generate and distribute animal models of disease to interested scientists — at the Wellcome Trust Sanger Institute. Male and female animals underwent a standardized phenotypic screen to determine the effects of deletion. Twenty six tests were carried out on mutant mice and two significant abnormalities were observed. No homozygous mutant embryos were recorded during gestation and, in a separate study, no homozygous animals were observed at weaning. The remaining tests were carried out on adult heterozygous mutant animals and no further abnormalities were observed. Interactions SNAPC4 has been shown to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SNAPC3
snRNA-activating protein complex subunit 3 is a protein that in humans is encoded by the ''SNAPC3'' gene. Interactions SNAPC3 has been shown to interact with SNAPC1 and retinoblastoma protein The retinoblastoma protein (protein name abbreviated pRb; gene name abbreviated ''Rb'', ''RB'' or ''RB1'') is a proto-oncogenic tumor suppressor protein that is dysfunctional in several major cancers. One function of pRb is to prevent excessive .... References Further reading * * * * * * * * {{Gene-9-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |