|
Ribonucleases
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalysis, catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 (for the phosphorolytic enzymes) and 3.1 (for the hydrolytic enzymes) classes of enzymes. Function All organisms studied contain many RNases of two different classes, showing that RNA degradation is a very ancient and important process. As well as clearing of cellular RNA that is no longer required, RNases play key roles in the maturation of all RNA molecules, both messenger RNAs that carry genetic material for making proteins and non-coding RNAs that function in varied cellular processes. In addition, active RNA degradation systems are the first defense against RNA viruses and provide the underlying machinery for more advanced cellular immune strategies such as RNAi. Some cells also secrete copious quantities of non-s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase H
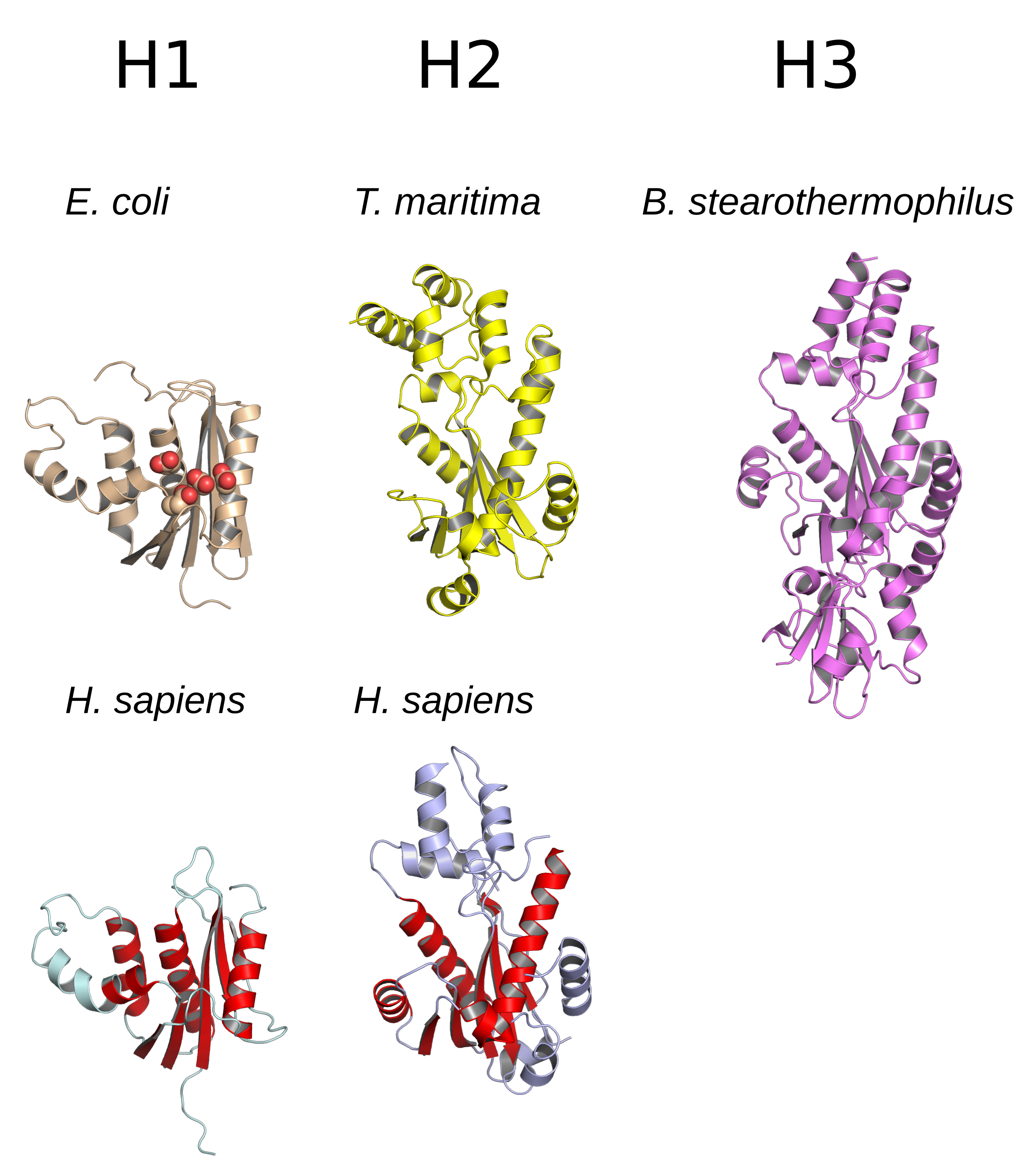
Ribonuclease H (abbreviated RNase H or RNH) is a family of non-nucleotide sequence, sequence-specific endonuclease enzymes that catalysis, catalyze the cleavage of RNA in an RNA/DNA substrate (chemistry), substrate via a hydrolysis, hydrolytic chemical mechanism, mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes. The family is divided into evolutionarily related groups with slightly different substrate (chemistry), substrate preferences, broadly designated ribonuclease H1 and H2. The human genome encodes both H1 and H2. Human ribonuclease H2 is a heterotrimeric complex composed of three subunits, mutations in any of which are among the genetic causes of a rare disease known as Aicardi–Goutières syndrome. A third type, closely related to H2, is found only in a few prokaryotes, whereas H1 and H2 occur in all domains of life. Additionally, RNase H1-like retroviral ribonuclease H domains occur in multidomain reve ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase PhyM
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 (for the phosphorolytic enzymes) and 3.1 (for the hydrolytic enzymes) classes of enzymes. Function All organisms studied contain many RNases of two different classes, showing that RNA degradation is a very ancient and important process. As well as clearing of cellular RNA that is no longer required, RNases play key roles in the maturation of all RNA molecules, both messenger RNAs that carry genetic material for making proteins and non-coding RNAs that function in varied cellular processes. In addition, active RNA degradation systems are the first defense against RNA viruses and provide the underlying machinery for more advanced cellular immune strategies such as RNAi. Some cells also secrete copious quantities of non-specific RN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase A
Pancreatic ribonuclease family (, ''RNase'', ''RNase I'', ''RNase A'', ''pancreatic RNase'', ''ribonuclease I'', ''endoribonuclease I'', ''ribonucleic phosphatase'', ''alkaline ribonuclease'', ''ribonuclease'', ''gene S glycoproteins'', ''Ceratitis capitata alkaline ribonuclease'', ''SLSG glycoproteins'', ''gene S locus-specific glycoproteins'', ''S-genotype-assocd. glycoproteins'', ''ribonucleate 3'-pyrimidino-oligonucleotidohydrolase'') is a superfamily of pyrimidine-specific endonucleases found in high quantity in the pancreas of certain mammals and of some reptiles. Specifically, the enzymes are involved in endonucleolytic cleavage of 3'-phosphomononucleotides and 3'-phosphooligonucleotides ending in C-P or U-P with 2',3'-cyclic phosphate intermediates. Ribonuclease can unwind the RNA helix by complexing with single-stranded RNA; the complex arises by an extended multi-site cation-anion interaction between lysine and arginine residues of the enzyme and phosphate groups of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase L
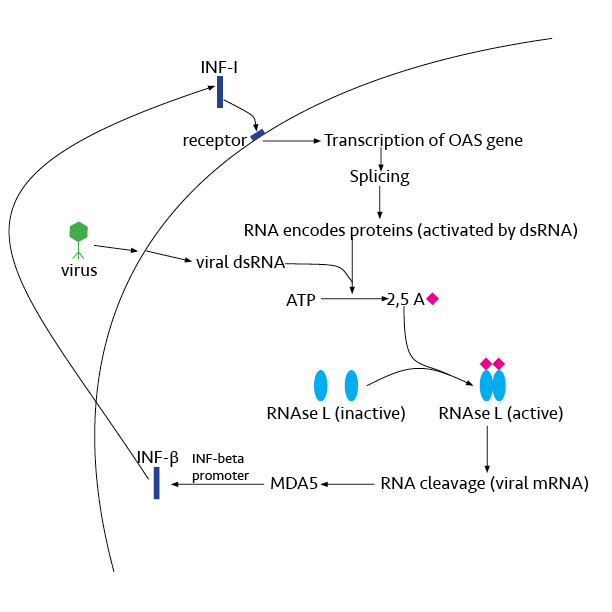
Ribonuclease L or RNase L (for ''latent''), known sometimes as ribonuclease 4 or 2'-5' oligoadenylate synthetase-dependent ribonuclease, is an interferon (IFN)-induced ribonuclease which, upon activation, destroys all RNA within the cell (both cellular and viral) as well as inhibiting mRNA export. RNase L is an enzyme that in humans is encoded by the ''RNASEL'' gene. This gene encodes a component of the interferon-regulated 2'-5'oligoadenylate (2'-5'A) system that functions in the antiviral and antiproliferative roles of interferons. RNase L is activated by dimerization, which occurs upon 2'-5'A binding, and results in cleavage of all RNA in the cell. This can lead to activation of MDA5, an RNA helicase involved in the production of interferons. Synthesis and activation RNase L is present in very minute quantities during the normal cell cycle. When interferon binds to cell receptors, it activates transcription of around 300 genes to bring about the antiviral state. Among ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase III
Ribonuclease III (RNase III or RNase C) (BREND3.1.26.3 is a type of ribonuclease that recognizes dsRNA and cleaves it at specific targeted locations to transform them into mature RNAs. These enzymes are a group of endoribonucleases that are characterized by their ribonuclease domain, which is labelled the RNase III domain. They are ubiquitous compounds in the cell and play a major role in pathways such as RNA precursor synthesis, RNA Silencing, and the ''pnp'' autoregulatory mechanism. Types of RNase III The RNase III superfamily is divided into four known classes: 1, 2, 3, and 4. Each class is defined by its domain structure.Liang Y-H, Lavoie M, Comeau M-A, Elela SA, Ji X. Structure of a Eukaryotic RNase III Post-Cleavage Complex Reveals a Double- Ruler Mechanism for Substrate Selection. Molecular cell. 2014;54(3):431-444. doi:10.1016/j.molcel.2014.03.006. Class 1 RNase III *Class 1 RNase III enzymes have a homodimeric structure whose function is to cleave dsRNA into multipl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase P
Ribonuclease P (, ''RNase P'') is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein-based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA molecules. Further, RNase P is one of two known multiple turnover ribozymes in nature (the other being the ribosome), the discovery of which earned Sidney Altman and Thomas Cech the Nobel Prize in Chemistry in 1989: in the 1970s, Altman discovered the existence of precursor tRNA with flanking sequences and was the first to characterize RNase P and its activity in processing of the 5' leader sequence of precursor tRNA. Its best characterised enzyme activity is the generation of mature 5′-ends of tRNAs by cleaving the 5′-leader elements of precursor-tRNAs. Cellular RNase Ps are ribonucleoproteins. The RNA from bacterial RNase P retains its catalytic activity in the ab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase T1
Ribonuclease T1 (, ''guanyloribonuclease'', ''Aspergillus oryzae ribonuclease'', ''RNase N1'', ''RNase N2'', ''ribonuclease N3'', ''ribonuclease U1'', ''ribonuclease F1'', ''ribonuclease Ch'', ''ribonuclease PP1'', ''ribonuclease SA'', ''RNase F1'', ''ribonuclease C2'', ''binase'', ''RNase Sa'', ''guanyl-specific RNase'', ''RNase G'', ''RNase T1'', ''ribonuclease guaninenucleotido-2'-transferase (cyclizing)'', ''ribonuclease N3'', ''ribonuclease N1'') is a fungal endonuclease that cleaves single-stranded RNA after guanine residues, i.e., on their 3' end; the most commonly studied form of this enzyme is the version found in the mold ''Aspergillus oryzae''. Owing to its specificity for guanine, RNase T1 is often used to digest denatured RNA prior to sequencing. Similar to other ribonucleases such as barnase and RNase A, ribonuclease T1 has been popular for folding studies. Structurally, ribonuclease T1 is a small α+β protein (104 amino acids) with a four-stranded, antiparall ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exoribonuclease
An exoribonuclease is an exonuclease ribonuclease, which are enzymes that degrade RNA by removing terminal nucleotides from either the 5' end or the 3' end of the RNA molecule. Enzymes that remove nucleotides from the 5' end are called ''5'-3' exoribonucleases'', and enzymes that remove nucleotides from the 3' end are called ''3'-5' exoribonucleases''. Exoribonucleases can use either water to cleave the nucleotide-nucleotide bond (which is called hydrolytic activity) or inorganic phosphate (which is called phosphorolytic activity). Hydrolytic exoribonucleases are classified under EC number 3.1 and phosphorolytic exoribonucleases under EC number 2.7.7. As the phosphorolytic enzymes use inorganic phosphate to cleave bonds they release nucleotide diphosphates, whereas the hydrolytic enzymes (which use water) release nucleotide monosphosphates. Exoribonucleases exist in all kingdoms of life, the bacteria, archaea, and eukaryotes. Exoribonucleases are involved in the degradati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribonuclease Inhibitor
Ribonuclease inhibitor (RI) is a large (~450 residues, ~49 kDa), acidic (pI ~4.7), leucine-rich repeat protein that forms extremely tight complexes with certain ribonucleases. It is a major cellular protein, comprising ~0.1% of all cellular protein by weight, and appears to play an important role in regulating the lifetime of RNA. RI has a surprisingly high cysteine content (~6.5%, cf. 1.7% in typical proteins) and is sensitive to oxidation. RI is also rich in leucine (21.5%, compared to 9% in typical proteins) and commensurately lower in other hydrophobic residues, esp. valine, isoleucine, methionine, tyrosine, and phenylalanine. Structure RI is the classic leucine-rich repeat protein, consisting of alternating alpha helix, α-helices and beta sheet, β-strands along its backbone. These secondary structure elements wrap around in a curved, right-handed solenoid that resembles a horseshoe. The parallel β-strands and α-helices form the inner and outer wall of the horseshoe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endoribonuclease
In biochemistry, an endoribonuclease is a class of enzyme which is a type of ribonuclease (an RNA cleaver), itself a type of endonuclease (a nucleotide cleaver). It cleaves either single-stranded or double-stranded RNA, depending on the enzyme. Example includes both single proteins such as RNase III, RNase A, RNase T1, RNase T2 and RNase H and also complexes of proteins with RNA such as RNase P and the RNA-induced silencing complex. Further examples include endoribonuclease XendoU found in frogs (''Xenopus ''Xenopus'' () (Gk., ξενος, ''xenos'' = strange, πους, ''pous'' = foot, commonly known as the clawed frog) is a genus of highly aquatic frogs native to sub-Saharan Africa. Twenty species are currently described with ...''). External links * {{Nucleases EC 3.1 Ribonucleases ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TRNA
Transfer ribonucleic acid (tRNA), formerly referred to as soluble ribonucleic acid (sRNA), is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes). In a cell, it provides the physical link between the genetic code in messenger RNA (mRNA) and the amino acid sequence of proteins, carrying the correct sequence of amino acids to be combined by the protein-synthesizing machinery, the ribosome. Each three-nucleotide codon in mRNA is complemented by a three-nucleotide anticodon in tRNA. As such, tRNAs are a necessary component of translation, the biological synthesis of new proteins in accordance with the genetic code. Overview The process of translation starts with the information stored in the nucleotide sequence of DNA. This is first transformed into mRNA, then tRNA specifies which three-nucleotide codon from the genetic code corresponds to which amino acid. Each mRNA codon is recognized by a particular type of tRNA, which docks to it along ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |