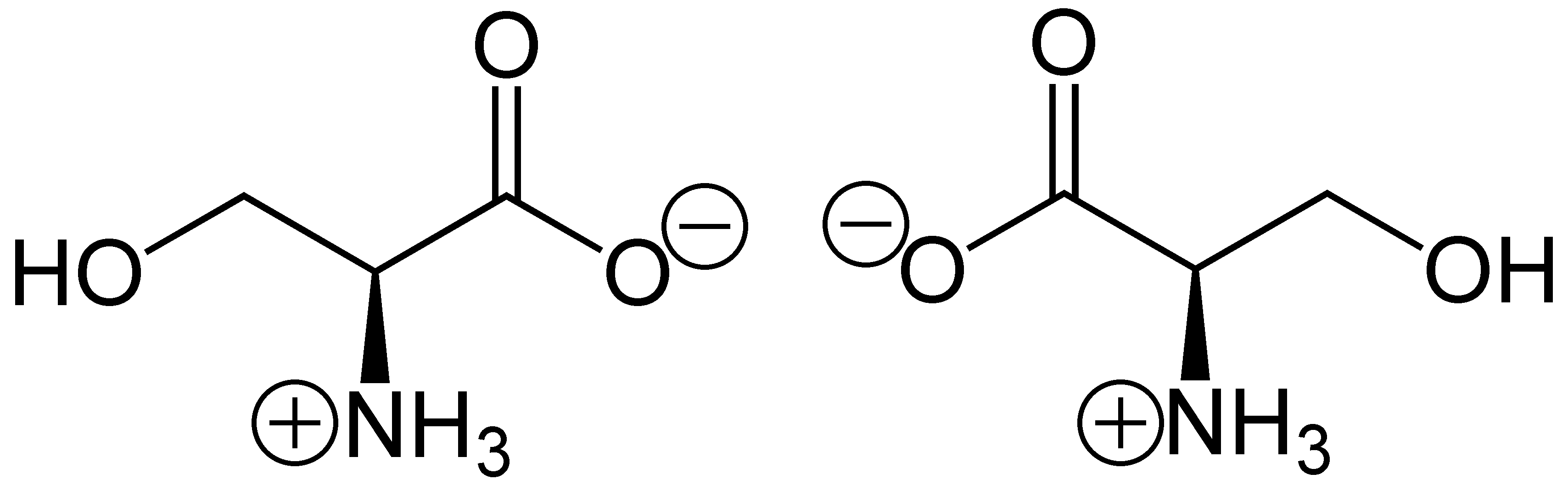|
Potyvirus
''Potyvirus'' is a genus of positive-strand RNA viruses in the family ''Potyviridae''. Plants serve as natural hosts. The genus is named after member virus ''potato virus Y''. Potyviruses account for about thirty percent of the currently known plant viruses. Like begomoviruses, members of this genus may cause significant losses in agricultural, pastoral, horticultural, and ornamental crops. More than 200 species of aphids spread potyviruses, and most are from the subfamily '' Aphidinae'' (genera ''Macrosiphum'' and ''Myzus''). The genus contains 190 species. Virology Structure The virion is non-enveloped with a flexuous and filamentous nucleocapsid, 680 to 900 nanometers (nm) long and is 11–20 nm in diameter. The nucleocapsid contains around 2000 copies of the capsid protein. The symmetry of the nucleocapsid is helical with a pitch of 3.4 nm. Genome The genome is a linear, positive-sense, single-stranded RNA ranging in size from 9,000–12,000 nucleoti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Potyviridae
''Potyviridae'' is a family of positive-strand RNA viruses that encompasses more than 30% of known plant viruses, many of which are of great agricultural significance. The family has 12 genera and 235 species, three of which are unassigned to a genus. Structure Potyvirid virions are nonenveloped, flexuous filamentous, rod-shaped particles. The diameter is around 12–15 nm, with a length of 200–300 nm. Genome Genomes are linear and usually nonsegmented, around 8–12kb in length, consisting of positive-sense RNA, which is surrounded by a protein coat made up of a single viral encoded protein called a capsid. All induce the formation of virus inclusion bodies called cylindrical inclusions (‘pinwheels’) in their hosts. These consist of a single protein (about 70 kDa) made in their hosts from a single viral genome product. Member viruses encode large polypeptides that are cleaved into mature proteins. In 5'–3' order these proteins are * P1 (a serine protea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Potato Virus Y
Potato virus Y (PVY) is a plant pathogenic virus of the family ''Potyviridae'', and one of the most important plant viruses affecting potato production. PVY infection of potato plants results in a variety of symptoms depending on the viral strain. The mildest of these symptoms is production loss, but the most detrimental is 'potato tuber necrotic ringspot disease' (PTNRD). Necrotic ringspots render potatoes unmarketable and can therefore result in a significant loss of income. PVY is transmissible by aphid vectors but may also remain dormant in seed potatoes. This means that using the same line of potato for production of seed potatoes for several consecutive generations will lead to a progressive increase in viral load and subsequent loss of crop. An increase in potato plant infection with viruses over the past few years has led to considerable losses to the South African potato industry. The increased rate of infection may be attributed to several factors. These include a ma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Helper-component Proteinase
Helper-component proteinase (, ''HC-Pro'') is an enzyme. This enzyme catalyses the following chemical reaction : Hydrolyses a Gly-Gly bond at its own C-terminus, commonly in the sequence -Tyr-Xaa-Val-Gly-Gly, in the processing of the potyviral polyprotein This enzyme is present in plant RNA viruses of the ''Potyviridae'' family. HC-Pro is encoded by all potyvirus ''Potyvirus'' is a genus of positive-strand RNA viruses in the family ''Potyviridae''. Plants serve as natural hosts. The genus is named after member virus ''potato virus Y''. Potyviruses account for about thirty percent of the currently known ...es, but is absent in some members of the '' Ipomovirus'' genus. HC-Pro is involved in virus transmission, virus polyprotein processing, and suppression of RNA silencing, an antiviral mechanism of plants. References External links * {{Portal bar, Biology, border=no EC 3.4.22 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plum Pox Virus
A plum is a fruit of some species in Prunus subg. Prunus, ''Prunus'' subg. ''Prunus''''.'' Dried plums are called prunes. History Plums may have been one of the first fruits domesticated by humans. Three of the most abundantly cultivated species are not found in the wild, only around human settlements: ''Prunus domestica'' has been traced to East European and Caucasian mountains, while ''Prunus salicina'' and ''Prunus simonii'' originated in China. Plum remains have been found in Neolithic age archaeological sites along with olives, grapes and figs. According to Ken Albala, plums originated in Iran. They were brought to Britain from Asia. An article on plum tree cultivation in Andalusia (southern Spain) appears in Ibn al-'Awwam's 12th-century agricultural work, ''Book on Agriculture''. Etymology and names The name plum derived from Old English ''plume'' "plum, plum tree", borrowed from Germanic language, Germanic or Middle Dutch, derived from Latin ' and ultimately from Anci ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sense (molecular Biology)
In molecular biology and genetics, the sense of a nucleic acid molecule, particularly of a strand of DNA or RNA, refers to the nature of the roles of the strand and its complement in specifying a sequence of amino acids. Depending on the context, sense may have slightly different meanings. For example, negative-sense strand of DNA is equivalent to the template strand, whereas the positive-sense strand is the non-template strand whose nucleotide sequence is equivalent to the sequence of the mRNA transcript. DNA sense Because of the complementary nature of base-pairing between nucleic acid polymers, a double-stranded DNA molecule will be composed of two strands with sequences that are reverse complements of each other. To help molecular biologists specifically identify each strand individually, the two strands are usually differentiated as the "sense" strand and the "antisense" strand. An individual strand of DNA is referred to as positive-sense (also positive (+) or simply sens ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Silencing
RNA silencing or RNA interference refers to a family of gene silencing effects by which gene expression is negatively regulated by non-coding RNAs such as microRNAs. RNA silencing may also be defined as sequence-specific regulation of gene expression triggered by double-stranded RNA (dsRNA). RNA silencing mechanisms are highly conserved in most eukaryotes. The most common and well-studied example is RNA interference (RNAi), in which endogenously expressed microRNA (miRNA) or exogenously derived small interfering RNA (siRNA) induces the degradation of complementary messenger RNA. Other classes of small RNA have been identified, including piwi-interacting RNA (piRNA) and its subspecies repeat associated small interfering RNA (rasiRNA). Background RNA silencing describes several mechanistically related pathways which are involved in controlling and regulating gene expression. RNA silencing pathways are associated with the regulatory activity of small non-coding RNAs (approximately 2 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryotic Initiation Factor 4F
Eukaryotic initiation factor 4F (eIF4F) is a heterotrimeric protein complex that binds the 5' cap of messenger RNAs (mRNAs) to promote eukaryotic translation initiation. The eIF4F complex is composed of three non-identical subunits: the DEAD-box RNA helicase eIF4A, the cap-binding protein eIF4E, and the large "scaffold" protein eIF4G. The mammalian eIF4F complex was first described in 1983, and has been a major area of study into the molecular mechanisms of cap-dependent translation initiation ever since. Function eIF4F is important for recruiting the small ribosomal subunit (40S) to the 5' cap of mRNAs during cap-dependent translation initiation. Components of the complex are also involved in cap-independent translation initiation; for instance, certain viral proteases cleave eIF4G to remove the eIF4E-binding region, thus inhibiting cap-dependent translation. Structure Structures of eIF4F components have been solved individually and as partial complexes by a variety o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-terminus
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus. Chemistry Each amino acid has a carboxyl group and an amine group. Amino acids link to one another to form a chain by a dehydration reaction which joins the amine group of one amino acid to the carboxyl group of the next. Thus polypeptide chains have an end with an unbound carboxyl group, the C-terminus, and an end with an unbound amine group, the N-terminus. Proteins are naturally synthesized starting from the N-terminus and ending at the C-terminus. Function C-terminal retention signals While the N-terminus of a protein often ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glycine
Glycine (symbol Gly or G; ) is an amino acid that has a single hydrogen atom as its side chain. It is the simplest stable amino acid ( carbamic acid is unstable), with the chemical formula NH2‐ CH2‐ COOH. Glycine is one of the proteinogenic amino acids. It is encoded by all the codons starting with GG (GGU, GGC, GGA, GGG). Glycine is integral to the formation of alpha-helices in secondary protein structure due to its compact form. For the same reason, it is the most abundant amino acid in collagen triple-helices. Glycine is also an inhibitory neurotransmitter – interference with its release within the spinal cord (such as during a ''Clostridium tetani'' infection) can cause spastic paralysis due to uninhibited muscle contraction. It is the only achiral proteinogenic amino acid. It can fit into hydrophilic or hydrophobic environments, due to its minimal side chain of only one hydrogen atom. History and etymology Glycine was discovered in 1820 by the French chemist ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protease
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalyzes (increases reaction rate or "speeds up") proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism (breakdown of old proteins), and cell signaling. In the absence of functional accelerants, proteolysis would be very slow, taking hundreds of years. Proteases can be found in all forms of life and viruses. They have independently evolved multiple times, and different classes of protease can perform the same reaction by completely different catalytic mechanisms. Hierarchy of proteases Based on catalytic residue Proteases can be classified into seven broad groups: * Serine protea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine
Serine (symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α- amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − form under biological conditions), and a side chain consisting of a hydroxymethyl group, classifying it as a polar amino acid. It can be synthesized in the human body under normal physiological circumstances, making it a nonessential amino acid. It is encoded by the codons UCU, UCC, UCA, UCG, AGU and AGC. Occurrence This compound is one of the naturally occurring proteinogenic amino acids. Only the L- stereoisomer appears naturally in proteins. It is not essential to the human diet, since it is synthesized in the body from other metabolites, including glycine. Serine was first obtained from silk protein, a particularly rich source, in 1865 by Emil Cramer. Its name is derived from the Latin for silk, ''sericum''. Serine's structure w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dalton (unit)
The dalton or unified atomic mass unit (symbols: Da or u) is a non-SI unit of mass widely used in physics and chemistry. It is defined as of the mass of an unbound neutral atom of carbon-12 in its nuclear and electronic ground state and at rest. The atomic mass constant, denoted ''m''u, is defined identically, giving . This unit is commonly used in physics and chemistry to express the mass of atomic-scale objects, such as atoms, molecules, and elementary particles, both for discrete instances and multiple types of ensemble averages. For example, an atom of helium-4 has a mass of . This is an intrinsic property of the isotope and all helium-4 atoms have the same mass. Acetylsalicylic acid (aspirin), , has an average mass of approximately . However, there are no acetylsalicylic acid molecules with this mass. The two most common masses of individual acetylsalicylic acid molecules are , having the most common isotopes, and , in which one carbon is carbon-13. The molecular ma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |