|
Pulse-chase Analysis
Pulse-chase analysis (PCA) is used to study the life cycles of proteins. Pulse-chase analysis experiments use radioactive and cytotoxic labels to "tag" proteins. Commonly used methods include treating cells with cycloheximide (CHX) to stop protein synthesis or radioisotopic amino acids or proteins such as green fluorescent protein (GFP). These labels are used to study proteins through their life cycles. While pulse-chase analysis is mainly used to study proteins, it can also be used to study different molecular structures that interact with proteins. Proteins can interact with different structures either because they are incorporated into the structure, such as in cells, or because they are part of a larger structure, such as in macromolecules. In biochemistry and molecular biology, a pulse-chase analysis is a method for examining a cellular process occurring over time by successively exposing the cells to a labeled compound (pulse) and then to the same compound in an unlabeled ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Folding
Protein folding is the physical process by which a protein, after Protein biosynthesis, synthesis by a ribosome as a linear chain of Amino acid, amino acids, changes from an unstable random coil into a more ordered protein tertiary structure, three-dimensional structure. This structure permits the protein to become biologically functional or active. The folding of many proteins begins even during the translation of the polypeptide chain. The amino acids interact with each other to produce a well-defined three-dimensional structure, known as the protein's native state. This structure is determined by the amino-acid sequence or primary structure. The correct three-dimensional structure is essential to function, although some parts of functional proteins Intrinsically unstructured proteins, may remain unfolded, indicating that protein dynamics are important. Failure to fold into a native structure generally produces inactive proteins, but in some instances, misfolded proteins have ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell Cycle
The cell cycle, or cell-division cycle, is the sequential series of events that take place in a cell (biology), cell that causes it to divide into two daughter cells. These events include the growth of the cell, duplication of its DNA (DNA replication) and some of its organelles, and subsequently the partitioning of its cytoplasm, chromosomes and other components into two daughter cells in a process called cell division. In eukaryotic cells (having a cell nucleus) including animal, plant, fungal, and protist cells, the cell cycle is divided into two main stages: interphase, and the M phase that includes mitosis and cytokinesis. During interphase, the cell grows, accumulating nutrients needed for mitosis, and replicates its DNA and some of its organelles. During the M phase, the replicated Chromosome, chromosomes, organelles, and cytoplasm separate into two new daughter cells. To ensure the proper replication of cellular components and division, there are control mechanisms kno ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
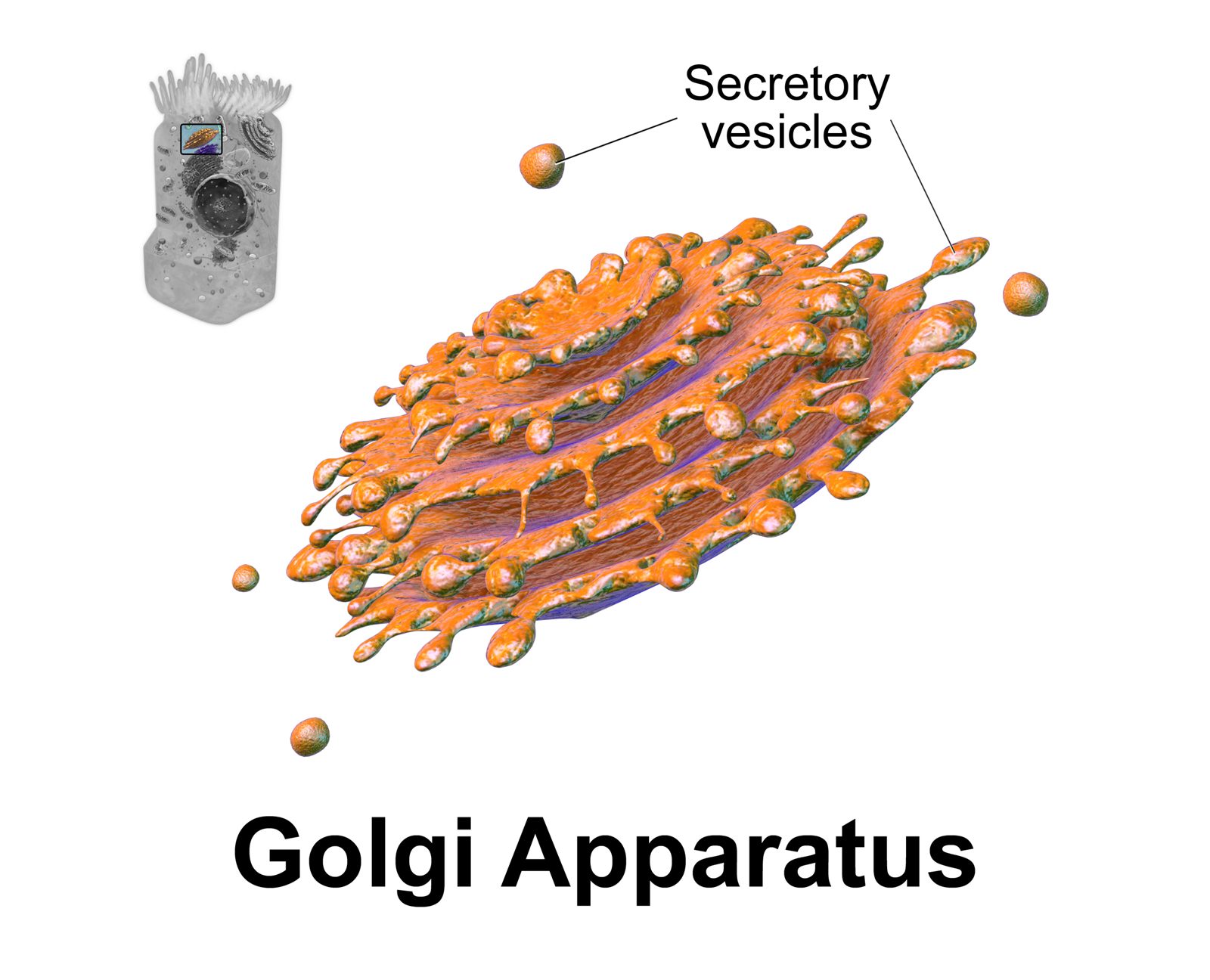
Golgi Apparatus
The Golgi apparatus (), also known as the Golgi complex, Golgi body, or simply the Golgi, is an organelle found in most eukaryotic Cell (biology), cells. Part of the endomembrane system in the cytoplasm, it protein targeting, packages proteins into membrane-bound Vesicle (biology and chemistry), vesicles inside the cell before the vesicles are sent to their destination. It resides at the intersection of the secretory, lysosomal, and Endocytosis, endocytic pathways. It is of particular importance in processing proteins for secretion, containing a set of glycosylation enzymes that attach various sugar monomers to proteins as the proteins move through the apparatus. The Golgi apparatus was identified in 1898 by the Italian biologist and pathologist Camillo Golgi. The organelle was later named after him in the 1910s. Discovery Because of its large size and distinctive structure, the Golgi apparatus was one of the first organelles to be discovered and observed in detail. It was d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Arabidopsis Thaliana
''Arabidopsis thaliana'', the thale cress, mouse-ear cress or arabidopsis, is a small plant from the mustard family (Brassicaceae), native to Eurasia and Africa. Commonly found along the shoulders of roads and in disturbed land, it is generally considered a weed. A winter annual with a relatively short lifecycle, ''A. thaliana'' is a popular model organism in plant biology and genetics. For a complex multicellular eukaryote, ''A. thaliana'' has a relatively small genome of around 135 Base pair#Length measurements, megabase pairs. It was the first plant to have its genome sequenced, and is an important tool for understanding the molecular biology of many plant traits, including flower development and phototropism, light sensing. Description ''Arabidopsis thaliana'' is an annual plant, annual (rarely biennial plant, biennial) plant, usually growing to 20–25 cm tall. The leaf, leaves form a rosette at the base of the plant, with a few leaves also on the flowering Plant ste ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hoechst Stain
Hoechst stains are part of a family of blue fluorescent dyes used to stain DNA. These bis-benzimides were originally developed by Hoechst AG, which numbered all their compounds so that the dye Hoechst 33342 is the 33,342nd compound made by the company. There are three related Hoechst stains: Hoechst 33258, Hoechst 33342, and Hoechst 34580. The dyes Hoechst 33258 and Hoechst 33342 are the ones most commonly used and they have similar excitation– emission spectra. Molecular characteristics Both dyes are excited by ultraviolet light at around 350 nm, and both emit blue-cyan fluorescent light around an emission spectrum maximum at 461 nm. Unbound dye has its maximum fluorescence emission in the 510–540 nm range. Hoechst stains can be excited with a xenon- or mercury-arc lamp or with an ultraviolet laser. There is a considerable Stokes shift between the excitation and emission spectra that makes Hoechst dyes useful in experiments in which multiple fluo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Propidium Iodide
Propidium iodide (or PI) is a Fluorescence#Biochemistry and medicine, fluorescent intercalating agent that can be used to Staining (biology), stain cell (biology), cells and nucleic acids. PI binds to DNA by intercalating between the bases with little or no sequence preference. When in an aqueous solution, PI has a fluorescent excitation maximum of 493 nm (blue-green), and an emission maximum of 636 nm (red). After binding DNA, the quantum yield of PI is enhanced 20-30 fold, and the excitation/emission maximum of PI is shifted to 535 nm (green) / 617 nm (orange-red). Propidium iodide is used as a DNA stain in flow cytometry to Viability assay, evaluate cell viability or DNA content in cell cycle analysis, or in microscopy to visualize the nucleus and other DNA-containing organelles. Propidium Iodide is not membrane-permeable, making it useful to differentiate Necrosis, necrotic, Apoptosis, apoptotic and healthy cells based on membrane integrity. PI also binds to RNA, necessitating ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DAPI
DAPI (pronounced 'DAPPY', /ˈdæpiː/), or 4′,6-diamidino-2-phenylindole, is a fluorescent stain that binds strongly to adenine– thymine-rich regions in DNA. It is used extensively in fluorescence microscopy. As DAPI can pass through an intact cell membrane, it can be used to stain both live and fixed cells, though it passes through the membrane less efficiently in live cells and therefore provides a marker for membrane viability. History DAPI was first synthesised in 1971 in the laboratory of Otto Dann as part of a search for drugs to treat trypanosomiasis. Although it was unsuccessful as a drug, further investigation indicated it bound strongly to DNA and became more fluorescent when bound. This led to its use in identifying mitochondrial DNA in ultracentrifugation in 1975, the first recorded use of DAPI as a fluorescent DNA stain. Strong fluorescence when bound to DNA led to the rapid adoption of DAPI for fluorescent staining of DNA for fluorescence microscopy. It ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Examples Of Markov Chains
Example may refer to: * ''exempli gratia'' (e.g.), usually read out in English as "for example" * .example, reserved as a domain name that may not be installed as a top-level domain of the Internet ** example.com, example.net, example.org, and example.edu: second-level domain names reserved for use in documentation as examples * HMS ''Example'' (P165), an Archer-class patrol and training vessel of the Royal Navy Arts * ''The Example'', a 1634 play by James Shirley * ''The Example'' (comics), a 2009 graphic novel by Tom Taylor and Colin Wilson * Example (musician), the British dance musician Elliot John Gleave (born 1982) * ''Example'' (album), a 1995 album by American rock band For Squirrels See also * Exemplar (other), a prototype or model which others can use to understand a topic better * Exemplum An exemplum (Latin for "example", exempla, ''exempli gratia'' = "for example", abbr.: ''e.g.'') is a moral anecdote, brief or extended, real or fictitious, us ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pri-miRNA
Micro ribonucleic acid (microRNA, miRNA, μRNA) are small, single-stranded, non-coding RNA molecules containing 21–23 nucleotides. Found in plants, animals, and even some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRNAs base-pair to complementary sequences in messenger RNA (mRNA) molecules, then silence said mRNA molecules by one or more of the following processes: * Cleaving the mRNA strand into two pieces. * Destabilizing the mRNA by shortening its poly(A) tail. * Reducing translation of the mRNA into proteins. In cells of humans and other animals, miRNAs primarily act by destabilizing the mRNA. miRNAs resemble the small interfering RNAs (siRNAs) of the RNA interference (RNAi) pathway, except miRNAs derive from regions of RNA transcripts that fold back on themselves to form short stem-loops (hairpins), whereas siRNAs derive from longer regions of double-stranded RNA. The human genome may encode over 1900 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MicroRNA
Micro ribonucleic acid (microRNA, miRNA, μRNA) are small, single-stranded, non-coding RNA molecules containing 21–23 nucleotides. Found in plants, animals, and even some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRNAs base-pair to complementary sequences in messenger RNA (mRNA) molecules, then silence said mRNA molecules by one or more of the following processes: * Cleaving the mRNA strand into two pieces. * Destabilizing the mRNA by shortening its poly(A) tail. * Reducing translation of the mRNA into proteins. In cells of humans and other animals, miRNAs primarily act by destabilizing the mRNA. miRNAs resemble the small interfering RNAs (siRNAs) of the RNA interference (RNAi) pathway, except miRNAs derive from regions of RNA transcripts that fold back on themselves to form short stem-loops (hairpins), whereas siRNAs derive from longer regions of double-stranded RNA. The human genome may encode ov ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Regulation
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products (protein or RNA). Sophisticated programs of gene expression are widely observed in biology, for example to trigger developmental pathways, respond to environmental stimuli, or adapt to new food sources. Virtually any step of gene expression can be modulated, from transcriptional initiation, to RNA processing, and to the post-translational modification of a protein. Often, one gene regulator controls another, and so on, in a gene regulatory network. Gene regulation is essential for viruses, prokaryotes and eukaryotes as it increases the versatility and adaptability of an organism by allowing the cell to express protein when needed. Although as early as 1951, Barbara McClintock showed interaction between two genetic loci, Activator (''Ac'') and Dissociator (''Ds''), in the color formation of maize seeds, t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |