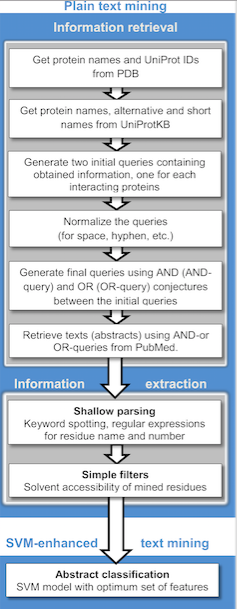
|
Proteome Analyst
Proteome Analyst (PA) is a freely available web server and online toolkit for predicting protein subcellular localization, or where a protein resides in a cell. In the field of proteomics, accurately predicting a protein's subcellular localization, or where a specific protein is located inside a cell, is an important step in the large scale study of proteins. This computational prediction problem is known as Protein subcellular localization prediction. Over the last decade, more than a dozen web servers and computer programs have been developed to attempt to solve this problem. Proteome Analyst is an example of one of the better performing subcellular prediction tools. Proteome Analyst makes predictions for both prokaryotic eukaryotic proteins using a text mining approach. Proteome Analyst was originally developed by the Proteome Analyst Research Group at the University of Alberta, and was initially released in March 2004. It was recently updated in January 2014. Input/Output and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
David S
David (; , "beloved one") was a king of ancient Israel and Judah and the Kings of Israel and Judah, third king of the Kingdom of Israel (united monarchy), United Monarchy, according to the Hebrew Bible and Old Testament. The Tel Dan stele, an Canaanite and Aramaic inscriptions, Aramaic-inscribed stone erected by a king of Aram-Damascus in the late 9th/early 8th centuries BCE to commemorate a victory over two enemy kings, contains the phrase (), which is translated as "Davidic line, House of David" by most scholars. The Mesha Stele, erected by King Mesha of Moab in the 9th century BCE, may also refer to the "House of David", although this is disputed. According to Jewish works such as the ''Seder Olam Rabbah'', ''Seder Olam Zutta'', and ''Sefer ha-Qabbalah'' (all written over a thousand years later), David ascended the throne as the king of Judah in 885 BCE. Apart from this, all that is known of David comes from biblical literature, Historicity of the Bible, the historicit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Subcellular Localization
The cells of eukaryotic organisms are elaborately subdivided into functionally-distinct membrane-bound compartments. Some major constituents of eukaryotic cells are: extracellular space, plasma membrane, cytoplasm, nucleus, mitochondria, Golgi apparatus, endoplasmic reticulum (ER), peroxisome, vacuoles, cytoskeleton, nucleoplasm, nucleolus, nuclear matrix and ribosomes. Bacteria also have subcellular localizations that can be separated when the cell is fractionated. The most common localizations referred to include the cytoplasm, the cytoplasmic membrane (also referred to as the inner membrane in Gram-negative bacteria), the cell wall (which is usually thicker in Gram-positive bacteria) and the extracellular environment. The cytoplasm, the cytoplasmic membrane and the cell wall are subcellular localizations, whereas the extracellular environment is clearly not. Most Gram-negative bacteria also contain an outer membrane and periplasmic space. Unlike eukaryotes, most bacteria c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteomics
Proteomics is the large-scale study of proteins. Proteins are vital macromolecules of all living organisms, with many functions such as the formation of structural fibers of muscle tissue, enzymatic digestion of food, or synthesis and replication of DNA. In addition, other kinds of proteins include antibodies that protect an organism from infection, and hormones that send important signals throughout the body. The proteome is the entire set of proteins produced or modified by an organism or system. Proteomics enables the identification of ever-increasing numbers of proteins. This varies with time and distinct requirements, or stresses, that a cell or organism undergoes. Proteomics is an interdisciplinary domain that has benefited greatly from the genetic information of various genome projects, including the Human Genome Project. It covers the exploration of proteomes from the overall level of protein composition, structure, and activity, and is an important component of function ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Subcellular Localization Prediction
Protein subcellular localization prediction (or just protein localization prediction) involves the prediction of where a protein resides in a cell, its subcellular localization. In general, prediction tools take as input information about a protein, such as a protein sequence of amino acids, and produce a predicted location within the cell as output, such as the nucleus, Endoplasmic reticulum, Golgi apparatus, extracellular space, or other organelles. The aim is to build tools that can accurately predict the outcome of protein targeting in cells. Prediction of protein subcellular localization is an important component of bioinformatics based prediction of protein function and genome annotation, and it can aid the identification of drug targets. Background Experimentally determining the subcellular localization of a protein can be a laborious and time consuming task. Immunolabeling or tagging (such as with a green fluorescent protein) to view localization using fluorescence m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Text Mining
Text mining, text data mining (TDM) or text analytics is the process of deriving high-quality information from text. It involves "the discovery by computer of new, previously unknown information, by automatically extracting information from different written resources." Written resources may include websites, books, emails, reviews, and articles. High-quality information is typically obtained by devising patterns and trends by means such as statistical pattern learning. According to Hotho et al. (2005), there are three perspectives of text mining: information extraction, data mining, and knowledge discovery in databases (KDD). Text mining usually involves the process of structuring the input text (usually parsing, along with the addition of some derived linguistic features and the removal of others, and subsequent insertion into a database), deriving patterns within the structured data, and finally evaluation and interpretation of the output. 'High quality' in text mining usually ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BLAST (biotechnology)
In bioinformatics, BLAST (basic local alignment search tool) is an algorithm and program for comparing Primary structure, primary biological sequence information, such as the amino acid, amino-acid sequences of proteins or the nucleotides of DNA sequence, DNA and/or RNA sequences. A BLAST search enables a researcher to compare a subject protein or nucleotide sequence (called a query) with a library or database of sequences, and identify database sequences that resemble the query sequence above a certain threshold. For example, following the discovery of a previously unknown gene in the Mus musculus, mouse, a scientist will typically perform a BLAST search of the human genome to see if humans carry a similar gene; BLAST will identify sequences in the human genome that resemble the mouse gene based on similarity of sequence. Background BLAST is one of the most widely used bioinformatics programs for sequence searching. It addresses a fundamental problem in bioinformatics research ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Uniprot
UniProt is a freely accessible database of protein sequence and functional information, many entries being derived from genome sequencing projects. It contains a large amount of information about the biological function of proteins derived from the research literature. It is maintained by the UniProt consortium, which consists of several European bioinformatics organisations and a foundation from Washington, DC, USA. The UniProt consortium The UniProt consortium comprises the European Bioinformatics Institute (EBI), the Swiss Institute of Bioinformatics (SIB), and the Protein Information Resource (PIR). EBI, located at the Wellcome Trust Genome Campus in Hinxton, UK, hosts a large resource of bioinformatics databases and services. SIB, located in Geneva, Switzerland, maintains the ExPASy (Expert Protein Analysis System) servers that are a central resource for proteomics tools and databases. PIR, hosted by the National Biomedical Research Foundation (NBRF) at the George ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Naive Bayes Classifier
In statistics, naive (sometimes simple or idiot's) Bayes classifiers are a family of " probabilistic classifiers" which assumes that the features are conditionally independent, given the target class. In other words, a naive Bayes model assumes the information about the class provided by each variable is unrelated to the information from the others, with no information shared between the predictors. The highly unrealistic nature of this assumption, called the naive independence assumption, is what gives the classifier its name. These classifiers are some of the simplest Bayesian network models. Naive Bayes classifiers generally perform worse than more advanced models like logistic regressions, especially at quantifying uncertainty (with naive Bayes models often producing wildly overconfident probabilities). However, they are highly scalable, requiring only one parameter for each feature or predictor in a learning problem. Maximum-likelihood training can be done by evaluating a c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fungi
A fungus (: fungi , , , or ; or funguses) is any member of the group of eukaryotic organisms that includes microorganisms such as yeasts and mold (fungus), molds, as well as the more familiar mushrooms. These organisms are classified as one of the kingdom (biology)#Six kingdoms (1998), traditional eukaryotic kingdoms, along with Animalia, Plantae, and either Protista or Protozoa and Chromista. A characteristic that places fungi in a different kingdom from plants, bacteria, and some protists is chitin in their cell walls. Fungi, like animals, are heterotrophs; they acquire their food by absorbing dissolved molecules, typically by secreting digestive enzymes into their environment. Fungi do not photosynthesize. Growth is their means of motility, mobility, except for spores (a few of which are flagellated), which may travel through the air or water. Fungi are the principal decomposers in ecological systems. These and other differences place fungi in a single group of related o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Prokaryotes
A prokaryote (; less commonly spelled procaryote) is a single-celled organism whose cell lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Ancient Greek (), meaning 'before', and (), meaning 'nut' or 'kernel'. In the earlier two-empire system arising from the work of Édouard Chatton, prokaryotes were classified within the empire Prokaryota. However, in the three-domain system, based upon molecular phylogenetics, prokaryotes are divided into two domains: Bacteria and Archaea. A third domain, Eukaryota, consists of organisms with nuclei. Prokaryotes evolved before eukaryotes, and lack nuclei, mitochondria, and most of the other distinct organelles that characterize the eukaryotic cell. Some unicellular prokaryotes, such as cyanobacteria, form colonies held together by biofilms, and large colonies can create multilayered microbial mats. Prokaryotes are asexual, reproducing via binary fission. Horizontal gene transfer is comm ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gram-positive
In bacteriology, gram-positive bacteria are bacteria that give a positive result in the Gram stain test, which is traditionally used to quickly classify bacteria into two broad categories according to their type of cell wall. The Gram stain is used by microbiologists to place bacteria into two main categories, gram-positive (+) and gram-negative bacteria, gram-negative (−). Gram-positive bacteria have a thick layer of peptidoglycan within the cell wall, and gram-negative bacteria have a thin layer of peptidoglycan. Gram-positive bacteria retain the crystal violet stain used in the test, resulting in a purple color when observed through an optical microscope. The thick layer of peptidoglycan in the bacterial cell wall retains the Stain (biology), stain after it has been fixed in place by iodine. During the decolorization step, the decolorizer removes crystal violet from all other cells. Conversely, gram-negative bacteria cannot retain the violet stain after the decolorization ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gram-negative
Gram-negative bacteria are bacteria that, unlike gram-positive bacteria, do not retain the crystal violet stain used in the Gram staining method of bacterial differentiation. Their defining characteristic is that their cell envelope consists of a thin peptidoglycan cell wall sandwiched between an inner ( cytoplasmic) membrane and an outer membrane. These bacteria are found in all environments that support life on Earth. Within this category, notable species include the model organism '' Escherichia coli'', along with various pathogenic bacteria, such as '' Pseudomonas aeruginosa'', '' Chlamydia trachomatis'', and '' Yersinia pestis''. They pose significant challenges in the medical field due to their outer membrane, which acts as a protective barrier against numerous antibiotics (including penicillin), detergents that would normally damage the inner cell membrane, and the antimicrobial enzyme lysozyme produced by animals as part of their innate immune system. Furthe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |