|
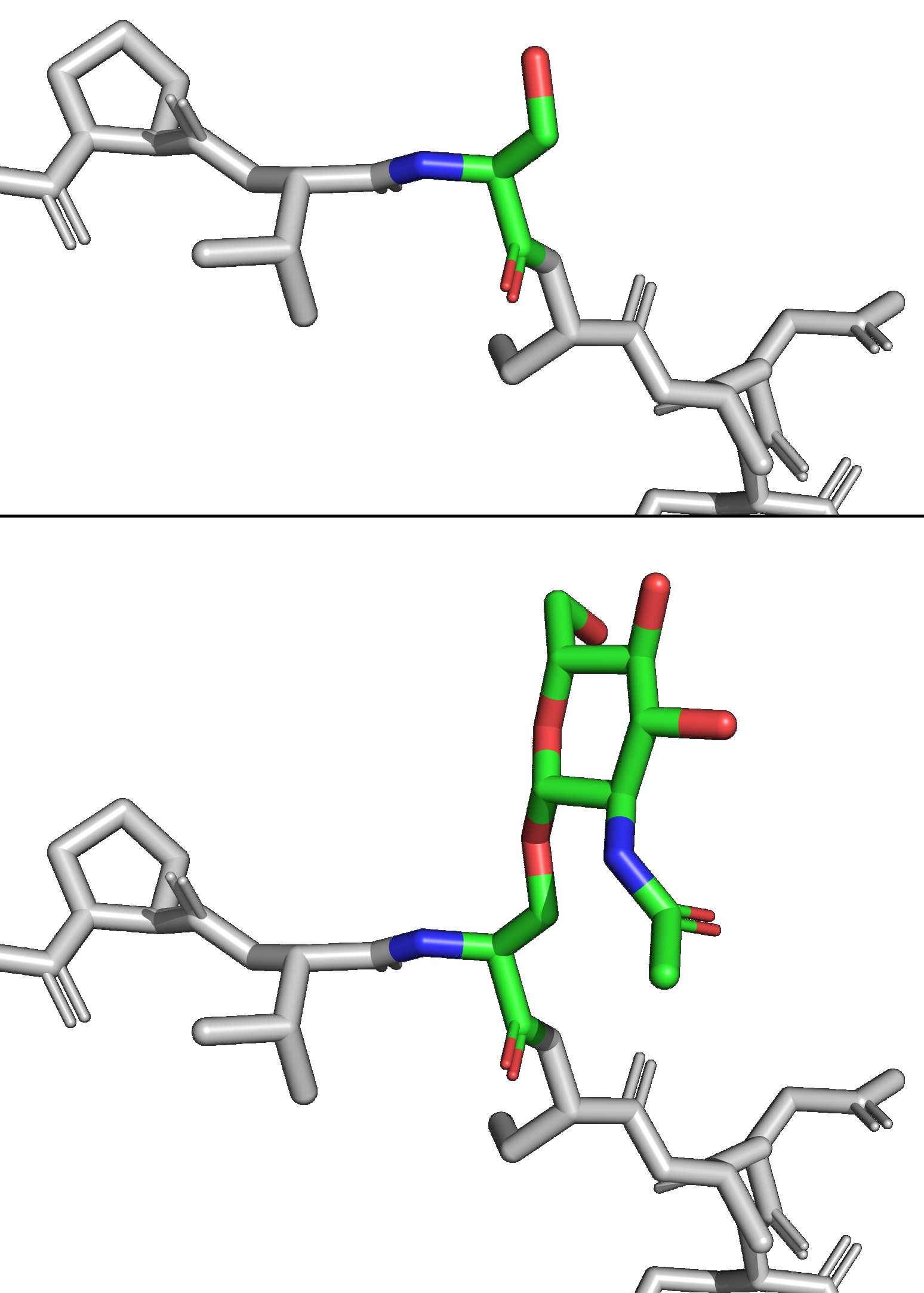
O-Linked β-N-acetylglucosamine
''O''-GlcNAc (short for ''O''-linked GlcNAc or ''O''-linked β-''N''-acetylglucosamine) is a reversible enzymatic post-translational modification that is found on serine and threonine residues of nucleocytoplasmic proteins. The modification is characterized by a β-glycosidic bond between the hydroxyl group of serine or threonine side chains and ''N''-acetylglucosamine (GlcNAc). ''O''-GlcNAc differs from other forms of protein glycosylation: (i) ''O''-GlcNAc is not elongated or modified to form more complex glycan structures, (ii) ''O''-GlcNAc is almost exclusively found on nuclear and cytoplasmic proteins rather than membrane proteins and secretory proteins, and (iii) ''O''-GlcNAc is a highly dynamic modification that turns over more rapidly than the proteins which it modifies. ''O''-GlcNAc is conserved across metazoans. Due to the dynamic nature of ''O''-GlcNAc and its presence on serine and threonine residues, ''O''-GlcNAcylation is similar to protein phosphorylation in so ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
O-GlcNAc Clear Red
''O''-GlcNAc (short for ''O''-linked GlcNAc or ''O''-linked β-''N''-acetylglucosamine) is a reversible Enzyme, enzymatic post-translational modification that is found on serine and threonine residues of Cell nucleus, nucleoCytoplasm, cytoplasmic proteins. The modification is characterized by a Glycosidic bond, β-glycosidic bond between the Hydroxy group, hydroxyl group of serine or threonine side chains and N-Acetylglucosamine, ''N''-acetylglucosamine (GlcNAc). ''O''-GlcNAc differs from other forms of protein glycosylation: (i) ''O''-GlcNAc is not elongated or modified to form more complex glycan structures, (ii) ''O''-GlcNAc is almost exclusively found on nuclear and cytoplasmic proteins rather than membrane proteins and secretory proteins, and (iii) ''O''-GlcNAc is a highly dynamic modification that turns over more rapidly than the proteins which it modifies. ''O''-GlcNAc is conserved across Animal, metazoans. Due to the dynamic nature of ''O''-GlcNAc and its presence on seri ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Phosphorylation
Protein phosphorylation is a reversible post-translational modification of proteins in which an amino acid residue is phosphorylated by a protein kinase by the addition of a covalently bound phosphate group. Phosphorylation alters the structural conformation of a protein, causing it to become activated, deactivated, or otherwise modifying its function. Approximately 13,000 human proteins have sites that are phosphorylated. The reverse reaction of phosphorylation is called dephosphorylation, and is catalyzed by protein phosphatases. Protein kinases and phosphatases work independently and in a balance to regulate the function of proteins. The amino acids most commonly phosphorylated are serine, threonine, tyrosine, and histidine. These phosphorylations play important and well-characterized roles in signaling pathways and metabolism. However, other amino acids can also be phosphorylated post-translationally, including arginine, lysine, aspartic acid, glutamic acid and cysteine, a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell Cycle
The cell cycle, or cell-division cycle, is the sequential series of events that take place in a cell (biology), cell that causes it to divide into two daughter cells. These events include the growth of the cell, duplication of its DNA (DNA replication) and some of its organelles, and subsequently the partitioning of its cytoplasm, chromosomes and other components into two daughter cells in a process called cell division. In eukaryotic cells (having a cell nucleus) including animal, plant, fungal, and protist cells, the cell cycle is divided into two main stages: interphase, and the M phase that includes mitosis and cytokinesis. During interphase, the cell grows, accumulating nutrients needed for mitosis, and replicates its DNA and some of its organelles. During the M phase, the replicated Chromosome, chromosomes, organelles, and cytoplasm separate into two new daughter cells. To ensure the proper replication of cellular components and division, there are control mechanisms kno ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Apoptosis
Apoptosis (from ) is a form of programmed cell death that occurs in multicellular organisms and in some eukaryotic, single-celled microorganisms such as yeast. Biochemistry, Biochemical events lead to characteristic cell changes (Morphology (biology), morphology) and death. These changes include Bleb (cell biology), blebbing, Plasmolysis, cell shrinkage, Karyorrhexis, nuclear fragmentation, Pyknosis, chromatin condensation, Apoptotic DNA fragmentation, DNA fragmentation, and mRNA decay. The average adult human loses 50 to 70 1,000,000,000, billion cells each day due to apoptosis. For the average human child between 8 and 14 years old, each day the approximate loss is 20 to 30 billion cells. In contrast to necrosis, which is a form of traumatic cell death that results from acute cellular injury, apoptosis is a highly regulated and controlled process that confers advantages during an organism's life cycle. For example, the separation of fingers and toes in a developing human embryo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenetics
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in addition to" the traditional (DNA sequence based) genetic mechanism of inheritance. Epigenetics usually involves a change that is not erased by cell division, and affects the regulation of gene expression. Such effects on cellular and physiological traits may result from environmental factors, or be part of normal development. The term also refers to the mechanism of changes: functionally relevant alterations to the genome that do not involve mutation of the nucleotide sequence. Examples of mechanisms that produce such changes are DNA methylation and histone modification, each of which alters how genes are expressed without altering the underlying DNA sequence. Further, non-coding RNA sequences have been shown to play a key role in the r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription (biology)
Transcription is the process of copying a segment of DNA into RNA for the purpose of gene expression. Some segments of DNA are transcribed into RNA molecules that can encode proteins, called messenger RNA (mRNA). Other segments of DNA are transcribed into RNA molecules called non-coding RNAs (ncRNAs). Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a Complementarity (molecular biology), complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, Antiparallel (biochemistry), antiparallel RNA strand called a primary transcript. In virology, the term transcription is used when referring to mRNA synthesis from a viral RNA molecule. The genome of many Orthornavirae, RNA viruses is composed of Sense (molecular biology), negative-sense RNA which acts as a template for positive sense viral messenger RNA - a necessary step in the synthesis of viral proteins needed for viral replication. This process ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteasome
Proteasomes are essential protein complexes responsible for the degradation of proteins by proteolysis, a chemical reaction that breaks peptide bonds. Enzymes that help such reactions are called proteases. Proteasomes are found inside all eukaryotes and archaea, and in some bacteria. In eukaryotes, proteasomes are located both in the nucleus and in the cytoplasm. The proteasomal degradation pathway is essential for many cellular processes, including the cell cycle, the regulation of gene expression, and responses to oxidative stress. The importance of proteolytic degradation inside cells and the role of ubiquitin in proteolytic pathways was acknowledged in the award of the 2004 Nobel Prize in Chemistry to Aaron Ciechanover, Avram Hershko and Irwin Rose. The core 20S proteasome (blue in the adjacent figure) is a cylindrical, compartmental protein complex of four stacked rings forming a central pore. Each ring is composed of seven individual proteins. The inner two rings a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Subcellular Localization
The cells of eukaryotic organisms are elaborately subdivided into functionally-distinct membrane-bound compartments. Some major constituents of eukaryotic cells are: extracellular space, plasma membrane, cytoplasm, nucleus, mitochondria, Golgi apparatus, endoplasmic reticulum (ER), peroxisome, vacuoles, cytoskeleton, nucleoplasm, nucleolus, nuclear matrix and ribosomes. Bacteria also have subcellular localizations that can be separated when the cell is fractionated. The most common localizations referred to include the cytoplasm, the cytoplasmic membrane (also referred to as the inner membrane in Gram-negative bacteria), the cell wall (which is usually thicker in Gram-positive bacteria) and the extracellular environment. The cytoplasm, the cytoplasmic membrane and the cell wall are subcellular localizations, whereas the extracellular environment is clearly not. Most Gram-negative bacteria also contain an outer membrane and periplasmic space. Unlike eukaryotes, most bacteria c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Structure
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, which are the monomers of the polymer. A single amino acid monomer may also be called a ''residue'', which indicates a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions, such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensiona ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein–protein Interaction
Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more protein molecules as a result of biochemical events steered by interactions that include electrostatic forces, hydrogen bonding and the hydrophobic effect. Many are physical contacts with molecular associations between chains that occur in a cell or in a living organism in a specific biomolecular context. Proteins rarely act alone as their functions tend to be regulated. Many molecular processes within a cell are carried out by molecular machines that are built from numerous protein components organized by their PPIs. These physiological interactions make up the so-called Interactome, interactomics of the organism, while aberrant PPIs are the basis of multiple aggregation-related diseases, such as Creutzfeldt–Jakob disease, Creutzfeldt–Jakob and Alzheimer's diseases. PPIs have been studied with Methods to investigate protein–protein interactions, many methods and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Uridine Diphosphate N-acetylglucosamine
Uridine diphosphate ''N''-acetylglucosamine or UDP-GlcNAc is a nucleotide sugar and a coenzyme in metabolism. It is used by glycosyltransferases to transfer ''N''-acetylglucosamine residues to substrates. UDP-GlcNAc is used for making glycosaminoglycans, proteoglycans, and glycolipids. D-Glucosamine is made naturally in the form of glucosamine-6-phosphate, and is the biochemical precursor of all nitrogen-containing sugars. To be specific, glucosamine-6-phosphate is synthesized from fructose 6-phosphate and glutamine as the first step of the hexosamine biosynthesis pathway. The end-product of this pathway is UDP-GlcNAc. Some enzymes involved in the biosynthesis of UDP-GlcNAc vary between prokaryotic and eukaryotic organisms, serving as potential drug targets for antibiotic development. Biosignaling UDP-GlcNAc is extensively involved in intracellular signaling as a substrate for ''O''-linked ''N''-acetylglucosamine transferases (OGTs) to install the ''O''-GlcNAc post-transl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein O-GlcNAcase
Protein ''O''-GlcNAcase (, OGA, glycoside hydrolase ''O''-GlcNAcase, ''O''-GlcNAcase, BtGH84, ''O''-GlcNAc hydrolase) is an enzyme with systematic name (protein)-3-''O''-(''N''-acetyl-D-glucosaminyl)-L-serine/threonine ''N''-acetylglucosaminyl hydrolase. OGA is encoded by the ''OGA'' gene. This enzyme catalyses the removal of the ''O''-GlcNAc post-translational modification in the following chemical reaction: # rotein3-''O''-(''N''-acetyl-β-D-glucosaminyl)-L-serine + H2O roteinL-serine + ''N''-acetyl-D-glucosamine # rotein3-''O''-(''N''-acetyl-β-D-glucosaminyl)-L-threonine + H2O roteinL-threonine + ''N''-acetyl-D-glucosamine Nomenclature Other names include: * Nuclear cytoplasmic ''O''-GlcNAcase and acetyltransferase Isoforms The human OGA gene is capable of producing two different transcripts, each capable of encoding a different OGA isoform. The long isoform L-OGA, a bifunctional enzyme that possess a glycoside hydrolase activity and a pseudo histone-acetyl tran ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |