|
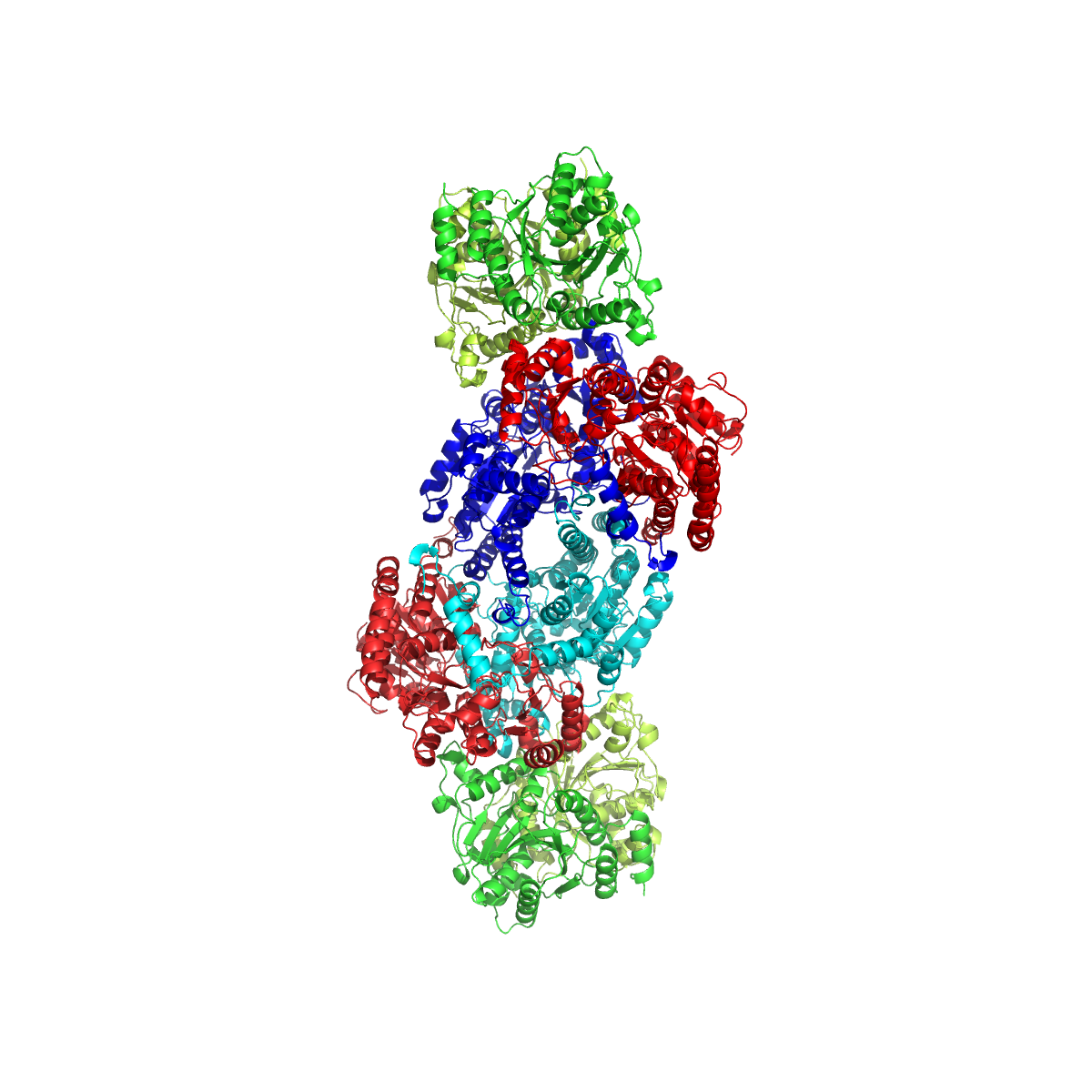
Nitrogenase
Nitrogenases are enzymes () that are produced by certain bacteria, such as cyanobacteria (blue-green bacteria) and rhizobacteria. These enzymes are responsible for the reduction of nitrogen (N2) to ammonia (NH3). Nitrogenases are the only family of enzymes known to catalyze this reaction, which is a key step in the process of nitrogen fixation. Nitrogen fixation is required for all forms of life, with nitrogen being essential for the biosynthesis of molecules (nucleotides, amino acids) that create plants, animals and other organisms. They are encoded by the Nif genes or homologs. They are related to protochlorophyllide reductase. Classification and structure Although the equilibrium formation of ammonia from molecular hydrogen and nitrogen has an overall negative enthalpy of reaction ( \Delta H^ = -45.2 \ \mathrm \, \mathrm \; \mathrm ), the activation energy is very high ( E_\mathrm = 230-420 \ \mathrm \, \mathrm ). Nitrogenase acts as a catalyst, reducing this energy ba ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nitrogenase
Nitrogenases are enzymes () that are produced by certain bacteria, such as cyanobacteria (blue-green bacteria) and rhizobacteria. These enzymes are responsible for the reduction of nitrogen (N2) to ammonia (NH3). Nitrogenases are the only family of enzymes known to catalyze this reaction, which is a key step in the process of nitrogen fixation. Nitrogen fixation is required for all forms of life, with nitrogen being essential for the biosynthesis of molecules (nucleotides, amino acids) that create plants, animals and other organisms. They are encoded by the Nif genes or homologs. They are related to protochlorophyllide reductase. Classification and structure Although the equilibrium formation of ammonia from molecular hydrogen and nitrogen has an overall negative enthalpy of reaction ( \Delta H^ = -45.2 \ \mathrm \, \mathrm \; \mathrm ), the activation energy is very high ( E_\mathrm = 230-420 \ \mathrm \, \mathrm ). Nitrogenase acts as a catalyst, reducing this energy ba ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nitrogen Fixation
Nitrogen fixation is a chemical process by which molecular nitrogen (), with a strong triple covalent bond, in the air is converted into ammonia () or related nitrogenous compounds, typically in soil or aquatic systems but also in industry. Atmospheric nitrogen is molecular dinitrogen, a relatively nonreactive molecule that is metabolically useless to all but a few microorganisms. Biological nitrogen fixation or ''diazotrophy'' is an important microbials mediated process that converts dinitrogen (N2) gas to ammonia (NH3) using the nitrogenase protein complex (Nif). Nitrogen fixation is essential to life because fixed inorganic nitrogen compounds are required for the biosynthesis of all nitrogen-containing organic compounds, such as amino acids and proteins, nucleoside triphosphates and nucleic acids. As part of the nitrogen cycle, it is essential for agriculture and the manufacture of fertilizer. It is also, indirectly, relevant to the manufacture of all nitrogen chemical ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FeMo Cofactor
FeMoco ( cofactor) is the primary Cofactor (biochemistry), cofactor of nitrogenase. Nitrogenase is the enzyme that catalyzes the conversion of atmospheric nitrogen molecules N2 into ammonia (NH3) through the process known as nitrogen fixation. Containing iron and molybdenum, the cofactor is called FeMoco. Its stoichiometry is Fe7MoS9C. Structure The FeMo cofactor is a Cluster chemistry, cluster with composition Fe7MoS9C. Fe is the Symbol (chemistry), chemical symbol for the element iron (ferrum), and Mo is the symbol for molybdenum. This large cluster can be viewed as two subunits composed of one Fe4S3 (iron(III) sulfide) cluster and one MoFe3S3 cluster. The two clusters are linked by three sulfide ligands. The unique iron (Fe) is anchored to the protein by a cysteine. It is also bound to three sulfides, resulting in tetrahedral molecular geometry. The additional six Fe centers in the cluster are each bonded to three sulfides. These six internal Fe centers define a trigonal p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FeMoco Cluster
FeMoco ( cofactor) is the primary Cofactor (biochemistry), cofactor of nitrogenase. Nitrogenase is the enzyme that catalyzes the conversion of atmospheric nitrogen molecules N2 into ammonia (NH3) through the process known as nitrogen fixation. Containing iron and molybdenum, the cofactor is called FeMoco. Its stoichiometry is Fe7MoS9C. Structure The FeMo cofactor is a Cluster chemistry, cluster with composition Fe7MoS9C. Fe is the Symbol (chemistry), chemical symbol for the element iron (ferrum), and Mo is the symbol for molybdenum. This large cluster can be viewed as two subunits composed of one Fe4S3 (iron(III) sulfide) cluster and one MoFe3S3 cluster. The two clusters are linked by three sulfide ligands. The unique iron (Fe) is anchored to the protein by a cysteine. It is also bound to three sulfides, resulting in tetrahedral molecular geometry. The additional six Fe centers in the cluster are each bonded to three sulfides. These six internal Fe centers define a trigonal p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ferredoxin
Ferredoxins (from Latin ''ferrum'': iron + redox, often abbreviated "fd") are iron–sulfur proteins that mediate electron transfer in a range of metabolic reactions. The term "ferredoxin" was coined by D.C. Wharton of the DuPont Co. and applied to the "iron protein" first purified in 1962 by Mortenson, Valentine, and Carnahan from the anaerobic bacterium '' Clostridium pasteurianum''. Another redox protein, isolated from spinach chloroplasts, was termed "chloroplast ferredoxin". The chloroplast ferredoxin is involved in both cyclic and non-cyclic photophosphorylation reactions of photosynthesis. In non-cyclic photophosphorylation, ferredoxin is the last electron acceptor thus reducing the enzyme NADP+ reductase. It accepts electrons produced from sunlight- excited chlorophyll and transfers them to the enzyme ferredoxin: NADP+ oxidoreductase . Ferredoxins are small proteins containing iron and sulfur atoms organized as iron–sulfur clusters. These biological "capacitors" can ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, Hot spring, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the nitrogen fixation, fixation of nitrogen from the Earth's atmosphere, atmosphere. The nutrient cycle includes the decomposition of cadaver, dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rhizobacteria
Rhizobacteria are root-associated bacteria that can have a detrimental (parasitic varieties), neutral or beneficial effect on plant growth. The name comes from the Greek ''rhiza'', meaning root. The term usually refers to bacteria that form symbiotic relationships with many plants ( mutualism). Rhizobacteria are often referred to as plant growth-promoting rhizobacteria, or PGPRs. The term PGPRs was first used by Joseph W. Kloepper in the late 1970s and has become commonly used in scientific literature. Generally, about 2–5% of rhizosphere bacteria are PGPR. They are an important group of microorganisms used in biofertilizer. Biofertilization accounts for about 65% of the nitrogen supply to crops worldwide. PGPRs have different relationships with different species of host plants. The two major classes of relationships are rhizospheric and endophytic. Rhizospheric relationships consist of the PGPRs that colonize the surface of the root, or superficial intercellular spaces of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ammonia
Ammonia is an inorganic compound of nitrogen and hydrogen with the formula . A stable binary hydride, and the simplest pnictogen hydride, ammonia is a colourless gas with a distinct pungent smell. Biologically, it is a common nitrogenous waste, particularly among aquatic organisms, and it contributes significantly to the nutritional needs of terrestrial organisms by serving as a precursor to 45% of the world's food and fertilizers. Around 70% of ammonia is used to make fertilisers in various forms and composition, such as urea and Diammonium phosphate. Ammonia in pure form is also applied directly into the soil. Ammonia, either directly or indirectly, is also a building block for the synthesis of many pharmaceutical products and is used in many commercial cleaning products. It is mainly collected by downward displacement of both air and water. Although common in nature—both terrestrially and in the outer planets of the Solar System—and in wide use, ammonia is bot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Iron–sulfur Cluster
Iron–sulfur clusters (or iron–sulphur clusters in British spelling) are molecular ensembles of iron and sulfide. They are most often discussed in the context of the biological role for iron–sulfur proteins, which are pervasive. Many Fe–S clusters are known in the area of organometallic chemistry and as precursors to synthetic analogues of the biological clusters (see Figure). It is believed that the last universal common ancestor had many iron-sulfur clusters. Organometallic clusters Organometallic Fe–S clusters include the sulfido carbonyls with the formula Fe2S2(CO)6, H2Fe3S(CO)9, and Fe3S2(CO)9. Compounds are also known that incorporate cyclopentadienyl ligands, such as (C5H5)4Fe4S4. Inorganic materials center, Structure of potassium dithioferrate, which features infinite chains of Fe(III) centers. Biological Fe–S clusters Iron–sulfur clusters occur in many biological systems, often as components of electron transfer proteins. The ferredoxin proteins a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenosine Triphosphate
Adenosine triphosphate (ATP) is an organic compound that provides energy to drive many processes in living cells, such as muscle contraction, nerve impulse propagation, condensate dissolution, and chemical synthesis. Found in all known forms of life, ATP is often referred to as the "molecular unit of currency" of intracellular energy transfer. When consumed in metabolic processes, it converts either to adenosine diphosphate (ADP) or to adenosine monophosphate (AMP). Other processes regenerate ATP. The human body recycles its own body weight equivalent in ATP each day. It is also a precursor to DNA and RNA, and is used as a coenzyme. From the perspective of biochemistry, ATP is classified as a nucleoside triphosphate, which indicates that it consists of three components: a nitrogenous base ( adenine), the sugar ribose, and the triphosphate. Structure ATP consists of an adenine attached by the 9-nitrogen atom to the 1′ carbon atom of a sugar ( ribose), which in tu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heterotetrameric
A tetrameric protein is a protein with a quaternary structure of four subunits (tetrameric). Homotetramers have four identical subunits (such as glutathione S-transferase), and heterotetramers are complexes of different subunits. A tetramer can be assembled as dimer of dimers with two homodimer subunits (such as sorbitol dehydrogenase), or two heterodimer subunits (such as hemoglobin). Subunit interactions in tetramers The interactions between subunits forming a tetramer is primarily determined by non covalent interaction. Hydrophobic effects, hydrogen bonds and electrostatic interactions are the primary sources for this binding process between subunits. For homotetrameric proteins such as Sorbitol dehydrogenase (SDH), the structure is believed to have evolved going from a monomeric to a dimeric and finally a tetrameric structure in evolution. The binding process in SDH and many other tetrameric enzymes can be described by the gain in free energy which can be determined ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
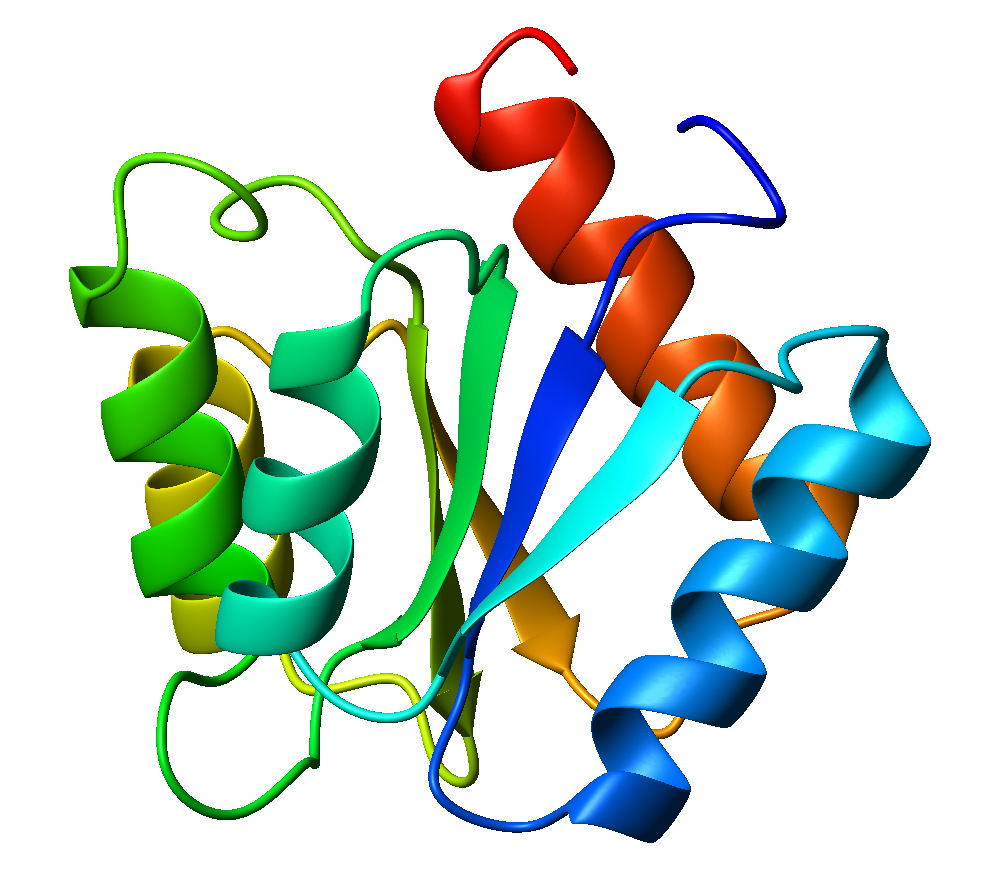
Flavodoxin
Flavodoxins (Fld) are small, soluble electron-transfer proteins. Flavodoxins contains flavin mononucleotide as prosthetic group. The structure of flavodoxin is characterized by a five-stranded parallel beta sheet, surrounded by five alpha helices. They have been isolated from prokaryotes, cyanobacteria, and some eukaryotic algae. Background Originally found in cyanobacteria and clostridia, flavodoxins were discovered over 50 years ago. These proteins evolved from an anaerobic environment, due to selective pressures. Ferredoxin, another redox protein, was the only protein able to be used in this manner. However, when oxygen became present in the environment, iron became limited. Ferredoxin is iron-dependant as well as oxidant-sensitive. Under these limited iron conditions, ferredoxin was no longer preferred. Flavodoxin on the other hand is the opposite of these traits, as it is oxidant-resistant and has iron-free isofunctional counterparts. Therefore, for some time flavodoxin was ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |

-3D-balls.png)