|
Mobilome
The mobilome is the entire set of mobile genetic elements in a genome. Mobilomes are found in eukaryotes, prokaryotes, and viruses. The compositions of mobilomes differ among lineages of life, with transposable elements being the major mobile elements in eukaryotes, and plasmids and prophages being the major types in prokaryotes. Virophages contribute to the viral mobilome. Mobilome in eukaryotes Transposable elements are elements that can move about or propagate within the genome, and are the major constituents of the eukaryotic mobilome. Transposable elements can be regarded as genetic parasites because they exploit the host cell's transcription and translation mechanisms to extract and insert themselves in different parts of the genome, regardless of the phenotypic effect on the host. Eukaryotic transposable elements were first discovered in maize (''Zea mays'') in which kernels showed a dotted color pattern. Barbara McClintock described the maize Ac/Ds system in whi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mobile Genetic Elements
Mobile genetic elements (MGEs), sometimes called selfish genetic elements, are a type of genetic material that can move around within a genome, or that can be transferred from one species or replicon to another. MGEs are found in all organisms. In humans, approximately 50% of the genome are thought to be MGEs. MGEs play a distinct role in evolution. Gene duplication events can also happen through the mechanism of MGEs. MGEs can also cause mutations in protein coding regions, which alters the protein functions. These mechanisms can also rearrange genes in the host genome generating variation. These mechanisms can increase fitness by gaining new or additional functions. An example of MGEs in evolutionary context are that virulence factors and antibiotic resistance genes of MGEs can be transported to share genetic code with neighboring bacteria. However, MGEs can also decrease fitness by introducing disease-causing alleles or mutations. The set of MGEs in an organism is called a mobil ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria and archaea; however plasmids are sometimes present in and eukaryotic organisms as well. Plasmids often carry useful genes, such as those involved in antibiotic resistance, virulence, secondary metabolism and bioremediation. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain additional genes for special circumstances. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the internet by various vendors ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Prophages
A prophage is a bacteriophage (often shortened to "phage") genome that is integrated into the circular bacterial chromosome or exists as an extrachromosomal plasmid within the bacterial cell. Integration of prophages into the bacterial host is the characteristic step of the lysogenic cycle of temperate phages. Prophages remain latent in the genome through multiple cell divisions until activation by an external factor, such as UV light, leading to production of new phage particles that will lyse the cell and spread. As ubiquitous mobile genetic elements, prophages play important roles in bacterial genetics and evolution, such as in the acquisition of virulence factors. Prophage induction Upon detection of host cell damage by UV light or certain chemicals, the prophage is excised from the bacterial chromosome in a process called prophage induction. After induction, viral replication begins via the lytic cycle. In the lytic cycle, the virus commandeers the cell's reproductive machi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reverse Transcriptase
A reverse transcriptase (RT) is an enzyme used to convert RNA genome to DNA, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, and by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes. The process does not violate the flows of genetic information as described by the classical central dogma, but rather expands it to include transfers of information from RNA to DNA. Retroviral RT has three sequential biochemical activities: RNA-dependent DNA polymerase activity, ribonuclease H (RNase H), and DNA-dependent DNA polymerase activity. Collectively, these activities enable the enzyme to convert single-stranded RNA into double-stranded cDNA. In retroviruses and retrotransposons, this cDNA can then integrate into the host genome, from which new RNA copies can be made via host-cell ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
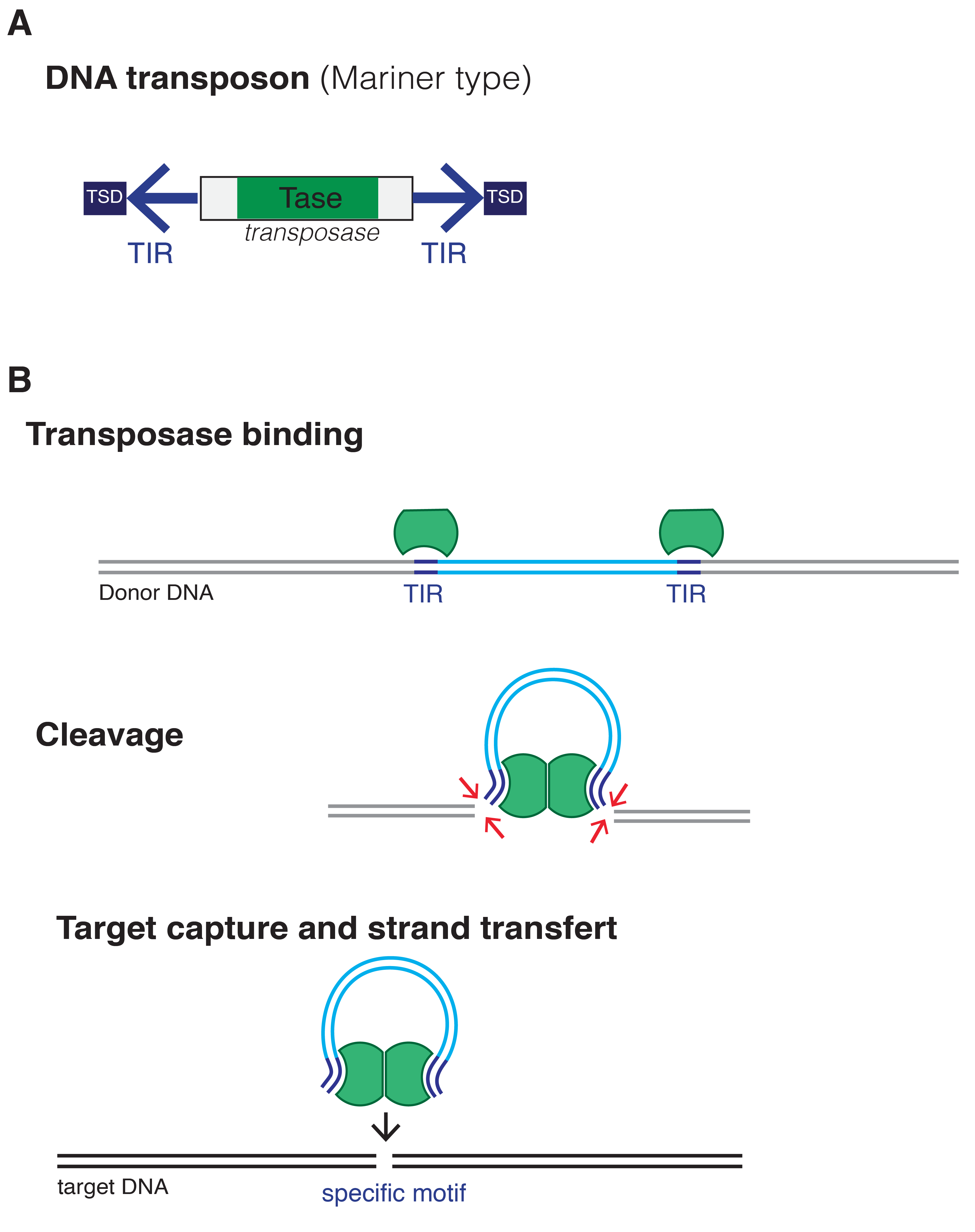
Retrotransposon
Retrotransposons (also called Class I transposable elements) are mobile elements which move in the host genome by converting their transcribed RNA into DNA through reverse transcription. Thus, they differ from Class II transposable elements, or DNA transposons, in utilizing an RNA intermediate for the transposition and leaving the transposition donor site unchanged. Through reverse transcription, retrotransposons amplify themselves quickly to become abundant in eukaryotic genomes such as maize (49–78%) and humans (42%). They are only present in eukaryotes but share features with retroviruses such as HIV, for example, discontinuous reverse transcriptase-mediated extrachromosomal recombination. There are two main types of retrotransposons, long terminal repeats (LTRs) and non-long terminal repeats (non-LTRs). Retrotransposons are classified based on sequence and method of transposition. Most retrotransposons in the maize genome are LTR, whereas in humans they are mostly non-L ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ty5 Retrotransposon
The Ty5 is a type of retrotransposon native to the ''Saccharomyces cerevisiae'' organism. The ''Saccharomyces cerevisiae'' retrotransposon Ty5 Ty5 is one of five endogenous retrotransposons native to the model organism ''Saccharomyces cerevisiae'', all of which target integration to gene poor regions. Endogenous retrotransposons are hypothesized to target gene poor chromosomal targets in order to reduce the chance of inactivating host genes. Ty1-Ty4 integrate upstream of Pol III promoters, while Ty5 targets integration to loci bound in heterochromatin. In the case of Ty5, this likely occurs by means of an interaction between the C-terminus of integrase and a target protein. The tight targeting patterns seen for the Ty elements are thought to be a means to limit damage to its host, which has a very gene dense genome. Ty5 was discovered in the mid 1990s in the laboratory of Daniel Voytas at Iowa State University. Ty5 is used as a model system by which to understand the biology of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Saccharomyces Cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungal microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have been originally isolated from the skin of grapes. It is one of the most intensively studied eukaryotic model organisms in molecular and cell biology, much like '' Escherichia coli'' as the model bacterium. It is the microorganism which causes many common types of fermentation. ''S. cerevisiae'' cells are round to ovoid, 5–10 μm in diameter. It reproduces by budding. Many proteins important in human biology were first discovered by studying their homologs in yeast; these proteins include cell cycle proteins, signaling proteins, and protein-processing enzymes. ''S. cerevisiae'' is currently the only yeast cell known to have Berkeley bodies present, which are involved in particular secretory pathways. Antibodies again ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Drosophila Melanogaster
''Drosophila melanogaster'' is a species of fly (an insect of the Order (biology), order Diptera) in the family Drosophilidae. The species is often referred to as the fruit fly or lesser fruit fly, or less commonly the "vinegar fly", "pomace fly", or "banana fly". In the wild, ''D. melanogaster'' are attracted to rotting fruit and fermenting beverages, and are often found in orchards, kitchens and pubs. Starting with Charles W. Woodworth's 1901 proposal of the use of this species as a model organism, ''D. melanogaster'' continues to be widely used for biological research in genetics, physiology, microbial pathogenesis, and Life history theory, life history evolution. ''D. melanogaster'' was the first animal to be Fruit flies in space, launched into space in 1947. As of 2017, six Nobel Prizes have been awarded to drosophilists for their work using the insect. ''Drosophila melanogaster'' is typically used in research owing to its rapid life cycle, relatively simple genetics with on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Barbara McClintock
Barbara McClintock (June 16, 1902 – September 2, 1992) was an American scientist and cytogenetics, cytogeneticist who was awarded the 1983 Nobel Prize in Physiology or Medicine. McClintock received her PhD in botany from Cornell University in 1927. There she started her career as the leader of the development of maize cytogenetics, the focus of her research for the rest of her life. From the late 1920s, McClintock studied chromosomes and how they change during reproduction in maize. She developed the technique for visualizing maize chromosomes and used microscopic analysis to demonstrate many fundamental genetic ideas. One of those ideas was the notion of genetic recombination by chromosomal crossover, crossing-over during meiosis—a mechanism by which chromosomes exchange information. She is often credited with producing the first genetic linkage, genetic map for maize, linking regions of the chromosome to physical traits. However this legend has been corrected by Kass (202 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coding Region
The coding region of a gene, also known as the coding DNA sequence (CDS), is the portion of a gene's DNA or RNA that codes for a protein. Studying the length, composition, regulation, splicing, structures, and functions of coding regions compared to non-coding regions over different species and time periods can provide a significant amount of important information regarding gene organization and evolution of prokaryotes and eukaryotes. This can further assist in mapping the human genome and developing gene therapy. Definition Although this term is also sometimes used interchangeably with exon, it is not the exact same thing: the exon can be composed of the coding region as well as the 3' and 5' untranslated regions of the RNA, and so therefore, an exon would be partially made up of coding region. The 3' and 5' untranslated regions of the RNA, which do not code for protein, are termed non-coding regions and are not discussed on this page. There is often confusion between coding ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Insertional Mutagenesis
In molecular biology, insertional mutagenesis is the creation of mutations in DNA by the addition of one or more base pairs. Such insertional mutations can occur naturally, mediated by viruses or transposons, or can be artificially created for research purposes in the lab. Signature tagged mutagenesis This is a technique used to study the function of genes. A transposon such as the P element of ''Drosophila melanogaster'' is allowed to integrate at random locations in the genome of the organism being studied. Mutants generated by this method are then screened for any unusual phenotypes. If such a phenotype is found then it can be assumed that the insertion has caused the gene relating to the usual phenotype to be inactivated. Because the sequence of the transposon is known, the gene can be identified, either by sequencing the whole genome and searching for the sequence, or by using the polymerase chain reaction to amplify specifically that gene. Virus insertional mutagenesis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biological Pigment
A biological pigment, also known simply as a pigment or biochrome, is a substance produced by living organisms that have a color resulting from selective Absorption (electromagnetic radiation), color absorption. Biological pigments include plant pigments and flower pigments. Many biological structures, such as skin, eyes, feathers, fur and hair contain pigments such as melanin in specialized cells called chromatophores. In some species, pigments accrue over very long periods during an individual's lifespan. Pigment color differs from structural color in that it is the same for all viewing angles, whereas structural color is the result of selective Reflection (physics), reflection or iridescence, usually because of multilayer structures. For example, butterfly wings typically contain structural color, although many butterflies have cells that contain pigment as well. Biological pigments See conjugated systems for electron bond chemistry that causes these molecules to have pigm ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |