|
Ligand Efficiency
Ligand efficiency is a measurement of the binding energy per atom of a ligand to its binding partner, such as a receptor or enzyme. Ligand efficiency is used in drug discovery research programs to assist in narrowing focus to lead compounds with optimal combinations of physicochemical properties and pharmacological properties. Mathematically, ligand efficiency (LE) can be defined as the ratio of Gibbs free energy (ΔG) to the number of non-hydrogen atoms of the compound: :LE = -(Δ''G'')/''N'' where Δ''G'' = −''RT''ln''Ki'' and ''N'' is the number of non-hydrogen atoms. It can be transformed to the equation: :LE = 1.4(−log IC50)/''N'' Other metrics Some suggest that better metrics for ligand efficiency are percentage/potency efficiency index (PEI), binding efficiency index (BEI) and surface-binding efficiency index (SEI) because they are easier to calculate and take into account the differences between elements in different rows of the periodic table. It is important t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Binding Energy
In physics and chemistry, binding energy is the smallest amount of energy required to remove a particle from a system of particles or to disassemble a system of particles into individual parts. In the former meaning the term is predominantly used in condensed matter physics, atomic physics, and chemistry, whereas in nuclear physics the term '' separation energy'' is used. A bound system is typically at a lower energy level than its unbound constituents. According to relativity theory, a decrease in the total energy of a system is accompanied by a decrease in the total mass, where . Types There are several types of binding energy, each operating over a different distance and energy scale. The smaller the size of a bound system, the higher its associated binding energy. Mass–energy relation A bound system is typically at a lower energy level than its unbound constituents because its mass must be less than the total mass of its unbound constituents. For systems with low bi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ligand
In coordination chemistry, a ligand is an ion or molecule with a functional group that binds to a central metal atom to form a coordination complex. The bonding with the metal generally involves formal donation of one or more of the ligand's electron pairs, often through Lewis acids and bases, Lewis bases. The nature of metal–ligand bonding can range from covalent bond, covalent to ionic bond, ionic. Furthermore, the metal–ligand bond order can range from one to three. Ligands are viewed as Lewis bases, although rare cases are known to involve Lewis acids and bases, Lewis acidic "ligands". Metals and metalloids are bound to ligands in almost all circumstances, although gaseous "naked" metal ions can be generated in a high vacuum. Ligands in a complex dictate the reactivity (chemistry), reactivity of the central atom, including ligand substitution rates, the reactivity of the ligands themselves, and redox. Ligand selection requires critical consideration in many practical are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
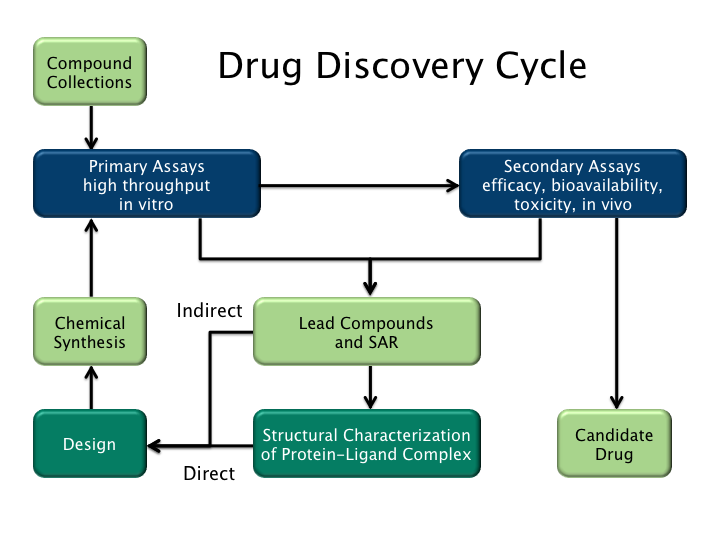
Drug Discovery
In the fields of medicine, biotechnology, and pharmacology, drug discovery is the process by which new candidate medications are discovered. Historically, drugs were discovered by identifying the active ingredient from traditional remedies or by serendipitous discovery, as with penicillin. More recently, chemical libraries of synthetic small molecules, natural products, or extracts were screened in intact cells or whole organisms to identify substances that had a desirable therapeutic effect in a process known as classical pharmacology. After sequencing of the human genome allowed rapid cloning and synthesis of large quantities of purified proteins, it has become common practice to use high throughput screening of large compounds libraries against isolated biological targets which are hypothesized to be disease-modifying in a process known as reverse pharmacology. Hits from these screens are then tested in cells and then in animals for efficacy. Modern drug discovery i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lead Compound
A lead compound (, i.e. a "leading" compound, not to be confused with various compounds of the metallic element lead) in drug discovery is a chemical compound that has pharmacological or biological activity likely to be therapeutically useful, but may nevertheless have suboptimal structure that requires modification to fit better to the target; lead drugs offer the prospect of being followed by back-up compounds. Its chemical structure serves as a starting point for chemical modifications in order to improve potency, selectivity, or pharmacokinetic parameters. Furthermore, newly invented pharmacologically active moieties may have poor druglikeness and may require chemical modification to become drug-like enough to be tested biologically or clinically. Terminology Lead compounds are sometimes called developmental candidates. This is because the discovery and selection of lead compounds occurs prior to preclinical and clinical development of the candidate. Discovering lead compo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gibbs Free Energy
In thermodynamics, the Gibbs free energy (or Gibbs energy as the recommended name; symbol is a thermodynamic potential that can be used to calculate the maximum amount of Work (thermodynamics), work, other than Work (thermodynamics)#Pressure–volume work, pressure–volume work, that may be performed by a closed system, thermodynamically closed system at constant temperature and pressure. It also provides a necessary condition for processes such as chemical reactions that may occur under these conditions. The Gibbs free energy is expressed as G(p,T) = U + pV - TS = H - TS where: * U is the internal energy of the system * H is the enthalpy of the system * S is the entropy of the system * T is the temperature of the system * V is the volume of the system * p is the pressure of the system (which must be equal to that of the surroundings for mechanical equilibrium). The Gibbs free energy change (, measured in joules in International System of Units, SI) is the ''maximum'' amount of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molar Concentration
Molar concentration (also called molarity, amount concentration or substance concentration) is the number of moles of solute per liter of solution. Specifically, It is a measure of the concentration of a chemical species, in particular, of a solute in a solution, in terms of amount of substance per unit volume of solution. In chemistry, the most commonly used unit for molarity is the number of moles per liter, having the unit symbol mol/L or mol/ dm3 (1000 mol/ m3) in SI units. A solution with a concentration of 1 mol/L is said to be 1 molar, commonly designated as 1 M or 1 M. Molarity is often depicted with square brackets around the substance of interest; for example, the molarity of the hydrogen ion is depicted as + Definition Molar concentration or molarity is most commonly expressed in units of moles of solute per litre of solution. For use in broader applications, it is defined as amount of substance of solute per unit volume of solution, or ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hit To Lead
Hit to lead (H2L) also known as lead generation is a stage in early drug discovery where small molecule hits from a high throughput screen (HTS) are evaluated and undergo limited optimization to identify promising lead compounds. These lead compounds undergo more extensive optimization in a subsequent step of drug discovery called lead optimization (LO). The drug discovery process generally follows the following path that includes a hit to lead stage: * Target validation (TV) → Assay development → High-throughput screening (HTS) → Hit to lead (H2L) → Lead optimization (LO) → Preclinical development → Clinical development The hit to lead stage starts with confirmation and evaluation of the initial screening hits and is followed by synthesis of analogs (hit expansion). Typically the initial screening hits display binding affinities for their biological target in the micromolar (10−6 molar concentration) range. Through limited H2L optimization, the affinities of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lipophilic Efficiency
Lipophilic efficiency (LiPE), sometimes referred to as ligand-lipophilicity efficiency (LLE) is a parameter used in drug design and drug discovery to evaluate the quality of research compounds, linking potency and lipophilicity in an attempt to estimate druglikeness. For a given compound LiPE is defined as the pIC50 (or pEC50) of interest minus the LogP of the compound. :\ce = \ce - \log P In practice, calculated values such as cLogP or calculated LogD are often used instead of the measured LogP or LogD. LiPE is used to compare compounds of different potencies (pIC50s) and lipophilicities (LogP). High potency (high value of pIC50) is a desirable attribute in drug candidates, as it reduces the risk of non-specific, off-target pharmacology at a given concentration. When associated with low clearance, high potency also allows for low total dose, which lowers the risk of idiosyncratic drug reaction. On the other hand, LogP is an estimate of a compound's overall lipophilicity, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Drug Design
Drug design, often referred to as rational drug design or simply rational design, is the invention, inventive process of finding new medications based on the knowledge of a biological target. The drug is most commonly an organic compound, organic small molecule that activates or inhibits the function of a biomolecule such as a protein, which in turn results in a therapeutic effect, therapeutic benefit to the patient. In the most basic sense, drug design involves the design of molecules that are complementary in shape and electric charge, charge to the biomolecular target with which they interact and therefore will bind to it. Drug design frequently but not necessarily relies on molecular modelling, computer modeling techniques. This type of modeling is sometimes referred to as computer-aided drug design. Finally, drug design that relies on the knowledge of the three-dimensional structure of the biomolecular target is known as structure-based drug design. In addition to small molec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Drug Discovery Hit To Lead
Hit to lead (H2L) also known as lead generation is a stage in early drug discovery where small molecule hits from a high-throughput screening, high throughput screen (HTS) are evaluated and undergo limited optimization to identify promising lead compounds. These lead compounds undergo more extensive optimization in a subsequent step of drug discovery called lead optimization (LO). The drug discovery process generally follows the following path that includes a hit to lead stage: * Target validation (TV) → Assay development → High-throughput screening (HTS) → Hit to lead (H2L) → #Lead optimization phase, Lead optimization (LO) → Preclinical development → Drug development#Clinical phase, Clinical development The hit to lead stage starts with confirmation and evaluation of the initial screening hits and is followed by synthesis of structural analog, analogs (hit expansion). Typically the initial screening hits display binding affinity, binding affinities for their biologic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Drug Discovery
In the fields of medicine, biotechnology, and pharmacology, drug discovery is the process by which new candidate medications are discovered. Historically, drugs were discovered by identifying the active ingredient from traditional remedies or by serendipitous discovery, as with penicillin. More recently, chemical libraries of synthetic small molecules, natural products, or extracts were screened in intact cells or whole organisms to identify substances that had a desirable therapeutic effect in a process known as classical pharmacology. After sequencing of the human genome allowed rapid cloning and synthesis of large quantities of purified proteins, it has become common practice to use high throughput screening of large compounds libraries against isolated biological targets which are hypothesized to be disease-modifying in a process known as reverse pharmacology. Hits from these screens are then tested in cells and then in animals for efficacy. Modern drug discovery i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |