|
H3K4me3
H3K4me3 is an epigenetic modification to the DNA packaging protein Histone H3 that indicates tri-methylation at the 4th lysine residue of the histone H3 protein and is often involved in the regulation of gene expression. The name denotes the addition of three methyl groups ( trimethylation) to the lysine 4 on the histone H3 protein. H3 is used to package DNA in eukaryotic cells (including human cells), and modifications to the histone alter the accessibility of genes for transcription. H3K4me3 is commonly associated with the activation of transcription of nearby genes. H3K4 trimethylation regulates gene expression through chromatin remodeling by the NURF complex. This makes the DNA in the chromatin more accessible for transcription factors, allowing the genes to be transcribed and expressed in the cell. More specifically, H3K4me3 is found to positively regulate transcription by bringing histone acetylases and nucleosome remodelling enzymes (NURF). H3K4me3 also plays an impor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone Methylation
Histone methylation is a process by which methyl groups are transferred to amino acids of histone proteins that make up nucleosomes, which the DNA double helix wraps around to form chromosomes. Methylation of histones can either increase or decrease transcription of genes, depending on which amino acids in the histones are methylated, and how many methyl groups are attached. Methylation events that weaken chemical attractions between histone tails and DNA increase transcription because they enable the DNA to uncoil from nucleosomes so that transcription factor proteins and RNA polymerase can access the DNA. This process is critical for the regulation of gene expression that allows different cells to express different genes. Function Histone methylation, as a mechanism for modifying chromatin structure is associated with stimulation of neural pathways known to be important for formation of long-term memories and learning. Animal models have shown methylation and other epigenetic re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
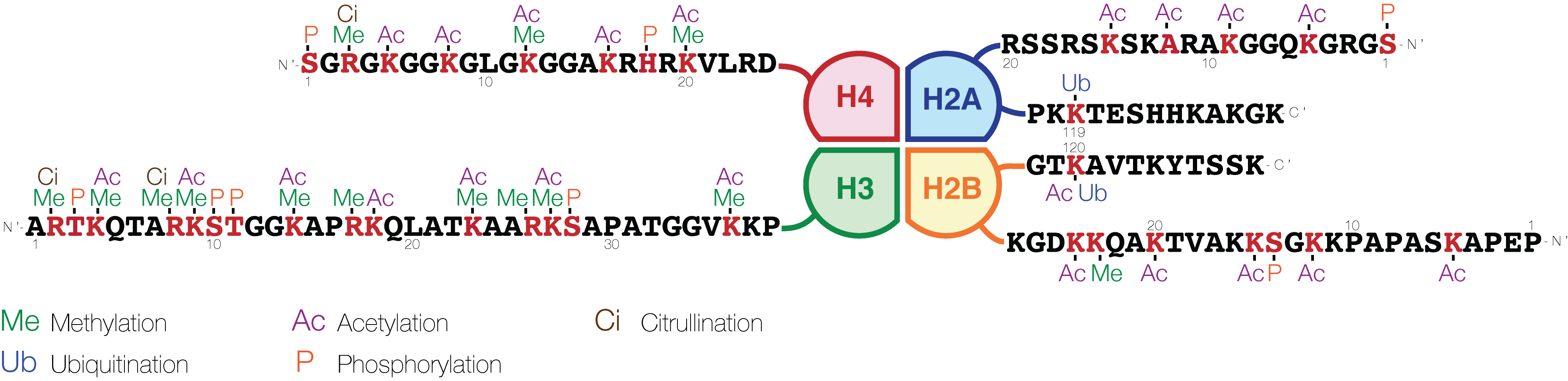
Histone Code
The histone code is a hypothesis that the transcription of genetic information encoded in DNA is in part regulated by chemical modifications (known as ''histone marks'') to histone proteins, primarily on their unstructured ends. Together with similar modifications such as DNA methylation it is part of the epigenetic code. Histones associate with DNA to form nucleosomes, which themselves bundle to form chromatin fibers, which in turn make up the more familiar chromosome. Histones are globular proteins with a flexible N-terminus (taken to be the tail) that protrudes from the nucleosome. Many of the histone tail modifications correlate very well to chromatin structure and both histone modification state and chromatin structure correlate well to gene expression levels. The critical concept of the histone code hypothesis is that the histone modifications serve to recruit other proteins by specific recognition of the modified histone via protein domains specialized for such purposes ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromatin Remodeling
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out by 1) covalent histone modifications by specific enzymes, e.g., histone acetyltransferases (HATs), deacetylases, methyltransferases, and kinases, and 2) ATP-dependent chromatin remodeling complexes which either move, eject or restructure nucleosomes. Besides actively regulating gene expression, dynamic remodeling of chromatin imparts an epigenetic regulatory role in several key biological processes, egg cells DNA replication and repair; apoptosis; chromosome segregation as well as development and pluripotency. Aberrations in chromatin remodeling proteins are found to be associated with human diseases, including cancer. Targeting chromatin remodeling pathways is currently evolving as a major therapeutic strategy in the treatment of several ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenetic
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in addition to" the traditional genetic basis for inheritance. Epigenetics most often involves changes that affect the regulation of gene expression, but the term can also be used to describe any heritable phenotypic change. Such effects on cellular and physiological phenotypic traits may result from external or environmental factors, or be part of normal development. The term also refers to the mechanism of changes: functionally relevant alterations to the genome that do not involve mutation of the nucleotide sequence. Examples of mechanisms that produce such changes are DNA methylation and histone modification, each of which alters how genes are expressed without altering the underlying DNA sequence. Gene expression can ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell Potency
Pluripotency: These are the cells that can generate into any of the three Germ layers which imply Endodermal, Mesodermal, and Ectodermal cells except tissues like the placenta. According to Latin terms, Pluripotentia means the ability for many things. We can generate Induced Pluripotent cells by using the Induced pluripotency technique by triggering or expressing the genes or the transcription factors of the normal somatic cells. They are abbreviated as iPSC or IPS. We can forcefully express the transcription factors like Oct4, Sox2, Klf4, and c-Myc of a non-pluripotent cell and convert them into a stem cell. This procedure is first studied in a Mouse fibroblast cell in 2006 and followed the same instructions in developing a Human pluripotent cell from a Human epidermal fibroblast cell. The technique is called Regeneration. Though the iPSC has similar properties to embryonic stem cells they were never approved for clinical stage research because they are highly Tumerogenic, hav ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Promoter
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Promoters control gene expression in bacteria and eukaryotes. RNA polymerase must attach to DNA near a gene for transcription to occur. Promoter DNA sequences provide an enzyme binding site. The -10 sequence is TATAAT. -35 sequences are conserved on average, but not in most promoters. Artificial promoters with conserved -10 and -35 elements transcribe more slowly. All D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) is a type of immunoprecipitation experimental technique used to investigate the interaction between proteins and DNA in the cell. It aims to determine whether specific proteins are associated with specific genomic regions, such as transcription factors on promoters or other DNA binding sites, and possibly define cistromes. ChIP also aims to determine the specific location in the genome that various histone modifications are associated with, indicating the target of the histone modifiers. ChIP is crucial for the advancements in the field of epigenomics and learning more about epigenetic phenomena. Briefly, the conventional method is as follows: # DNA and associated proteins on chromatin in living cells or tissues are crosslinked (this step is omitted in Native ChIP). # The DNA-protein complexes (chromatin-protein) are then sheared into ~500 bp DNA fragments by sonication or nuclease digestion. # Cross-linked DNA fragments associated with the prot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Start Site
Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1–3% of total RNA samples. Less than 2% of the human genome can be transcribed into mRNA ( Human genome#Coding vs. noncoding DNA), while at least 80% of mammalian genomic DNA can be actively transcribed (in one or more types of cells), with the majority of this 80% considered to be ncRNA. Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript. Transcription proceeds in the following general steps: # RNA polymerase, together with one or more general transcription factors, binds to promoter DNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hox Gene
Hox genes, a subset of homeobox genes, are a group of related genes that specify regions of the body plan of an embryo along the head-tail axis of animals. Hox proteins encode and specify the characteristics of 'position', ensuring that the correct structures form in the correct places of the body. For example, Hox genes in insects specify which appendages form on a segment (for example, legs, antennae, and wings in fruit flies), and Hox genes in vertebrates specify the types and shape of vertebrae that will form. In segmented animals, Hox proteins thus confer segmental or positional identity, but do not form the actual segments themselves. Studies on Hox genes in ciliated larvae have shown they are only expressed in future adult tissues. In larvae with gradual metamorphosis the Hox genes are activated in tissues of the larval body, generally in the trunk region, that will be maintained through metamorphosis. In larvae with complete metamorphosis the Hox genes are mainly express ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Structural Motif
In a polymer, chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common Biomolecular structure#Tertiary structure, three-dimensional structure that appears in a variety of different, evolutionarily unrelated molecules. A structural motif does not have to be associated with a sequence motif; it can be represented by different and completely unrelated sequences in different proteins or RNA. In nucleic acids Depending upon the sequence and other conditions, nucleic acids can form a variety of structural motifs which is thought to have biological significance. ;Stem-loop: Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
WD40 Repeat
The WD40 repeat (also known as the WD or beta-transducin repeat) is a short structural motif of approximately 40 amino acids, often terminating in a tryptophan-aspartic acid (W-D) dipeptide. Tandem copies of these repeats typically fold together to form a type of circular solenoid protein domain called the WD40 domain. Structure WD40 domain-containing proteins have 4 to 16 repeating units, all of which are thought to form a circularised beta-propeller structure (see figure to the right). The WD40 domain is composed of several repeats, a variable region of around 20 residues at the beginning followed by a more common repeated set of residues. These repeats typically form a four stranded anti-parallel beta sheet or blade. These blades come together to form a propeller with the most common being a 7 bladed beta propeller. The blades interlock so that the last beta strand of one repeat forms with the first three of the next repeat to form the 3D blade structure. Function WD40-repe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
WDR5
WD repeat-containing protein 5 is a protein that in humans is encoded by the ''WDR5'' gene. This gene encodes a member of the WD repeat protein family. WD repeats are minimally conserved regions of approximately 40 amino acids typically bracketed by gly-his and trp-asp (GH-WD), which may facilitate formation of heterotrimeric or multiprotein complexes. Members of this family are involved in a variety of cellular processes, including cell cycle progression, signal transduction, apoptosis, and gene regulation. This protein contains 7 WD repeats. Alternatively spliced transcript variants encoding the same protein have been identified. Interactions WDR5 has been shown to interact with Host cell factor C1 and MLL. It also interacts with the long non-coding RNA HOTTIP and to the lncRNA NeST. WDR5 is a key determinant for MYC recruitment to chromatin Chromatin is a complex of DNA and protein found in eukaryotic cells. The primary function is to package long DNA molecules into more ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |