|
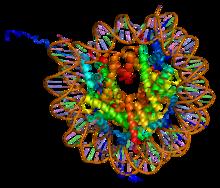
Histone Fold
The histone fold is a structural motif located near the C-terminus of histone proteins, characterized by three alpha helices separated by two loops. This motif facilitates the formation of heterodimers, which subsequently assemble into a histone octamer, playing a crucial role in the packaging of DNA into nucleosomes within chromatin. This fold is an ancient and highly conserved structural motif, essential for DNA compaction and regulation across a wide range of species. Discovery The histone fold motif was first discovered in TATA box-binding protein-associated factors, which play a key role in transcription. Structure The histone fold is typically around 70 amino acids long and is characterized by three alpha helices connected by two short, unstructured loops. In the absence of DNA, core histones assemble into head-to-tail intermediates. For instance, H3 and H4 first form heterodimers, which then combine to form a tetramer. Similarly, H2A and H2B form heterodimers. Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Structural Motif
In a chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common three-dimensional structure that appears in a variety of different, evolutionarily unrelated molecules. A structural motif does not have to be associated with a sequence motif; it can be represented by different and completely unrelated sequences in different proteins or RNA. In nucleic acids Depending upon the sequence and other conditions, nucleic acids can form a variety of structural motifs which is thought to have biological significance. ;Stem-loop: Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The resulting structure is a key building block of many ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H3
Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal end, N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'beads on a string' structure. Histone proteins are highly post-translationally modified however Histone H3 is the most extensively modified of the five histones. The term "Histone H3" alone is purposely ambiguous in that it does not distinguish between sequence variants or modification state. Histone H3 is an important protein in the emerging field of epigenetics, where its sequence variants and variable modification states are thought to play a role in the dynamic and long term regulation of genes. Epigenetics and post-translational modifications The N-terminus of H3 protrudes from the globular nucleosome core and is susceptible to post-translational modification that influence cellular processes. These modifications include the covalen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Folding
Protein folding is the physical process by which a protein, after Protein biosynthesis, synthesis by a ribosome as a linear chain of Amino acid, amino acids, changes from an unstable random coil into a more ordered protein tertiary structure, three-dimensional structure. This structure permits the protein to become biologically functional or active. The folding of many proteins begins even during the translation of the polypeptide chain. The amino acids interact with each other to produce a well-defined three-dimensional structure, known as the protein's native state. This structure is determined by the amino-acid sequence or primary structure. The correct three-dimensional structure is essential to function, although some parts of functional proteins Intrinsically unstructured proteins, may remain unfolded, indicating that protein dynamics are important. Failure to fold into a native structure generally produces inactive proteins, but in some instances, misfolded proteins have ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Drosophila
''Drosophila'' (), from Ancient Greek δρόσος (''drósos''), meaning "dew", and φίλος (''phílos''), meaning "loving", is a genus of fly, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or pomace flies, vinegar flies, or wine flies, a reference to the characteristic of many species to linger around overripe or rotting fruit. They should not be confused with the Tephritidae, a related family, which are also called fruit flies (sometimes referred to as "true fruit flies"); tephritids feed primarily on unripe or ripe fruit, with many species being regarded as destructive agricultural pests, especially the Mediterranean fruit fly. One species of ''Drosophila'' in particular, ''Drosophila melanogaster'', has been heavily used in research in genetics and is a common model organism in developmental biology. The terms "fruit fly" and "''Drosophila''" are often used synonymously with ''D. melanogaster'' in modern biological literatur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aquifex Aeolicus
''Aquifex'' is a bacterial genus, belonging to phylum Aquificota. There is one species of ''Aquifex'' with a validly published name – '' A. pyrophilus'' – but "'' A. aeolicus''" is sometimes considered as species though it has no standing as a name given it has not been validly or effectively published. ''Aquifex'' spp. are extreme thermophiles, growing best at temperature of 85 °C to 95 °C. They are members of the ''Bacteria'' as opposed to the other inhabitants of extreme environments, the Archaea. ''Aquifex'' spp. are rod-shaped bacteria with a length of 2 to 6 μm, have a diameter of around 0.5 μm and are motile. They are non-sporeforming, Gram negative autotrophs. ''Aquifex'' means ''water-maker'' in Latin, and refers to the fact that its method of respiration creates water. ''Aquifex'' tend to form cell aggregates composed of up to 100 individual cells. ''Aquifex'' spp. are thermophilic and often grow near underwater volcanoes or hot springs. ''A. aeo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptide
Peptides are short chains of amino acids linked by peptide bonds. A polypeptide is a longer, continuous, unbranched peptide chain. Polypeptides that have a molecular mass of 10,000 Da or more are called proteins. Chains of fewer than twenty amino acids are called oligopeptides, and include dipeptides, tripeptides, and tetrapeptides. Peptides fall under the broad chemical classes of biological polymers and oligomers, alongside nucleic acids, oligosaccharides, polysaccharides, and others. Proteins consist of one or more polypeptides arranged in a biologically functional way, often bound to ligands such as coenzymes and cofactors, to another protein or other macromolecule such as DNA or RNA, or to complex macromolecular assemblies. Amino acids that have been incorporated into peptides are termed residues. A water molecule is released during formation of each amide bond.. All peptides except cyclic peptides have an N-terminal (amine group) and C-terminal (carboxyl g ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Acetylated
: In chemistry, acetylation is an organic esterification reaction with acetic acid. It introduces an acetyl group into a chemical compound. Such compounds are termed ''acetate esters'' or simply ''acetates''. Deacetylation is the opposite reaction, the removal of an acetyl group from a chemical compound. Acetylation/deacetylation in biology Histone deacetylases "play crucial roles in gene transcription and most likely in all eukaryotic biological processes that involve chromatin". Acetylation is one type of post-translational modification of proteins. The acetylation of the ε-amino group of lysine, which is common, converts a charged side chain to a neutral one. Acetylation/deacetylation of histones also plays a role in gene expression and cancer. These modifications are effected by enzymes called histone acetyltransferases (HATs) and histone deacetylases (HDACs). Two general mechanisms are known for deacetylation. One mechanism involves zinc binding to the acetyl oxyg ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H2B
Histone H2B is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and long N-terminal and C-terminal tails, H2B is involved with the structure of the nucleosomes. Structure Histone H2B is a lightweight structural protein made of 126 amino acids. Many of these amino acids have a positive charge at cellular pH, which allows them to interact with the negatively charged phosphate groups in DNA. Along with a central globular domain, histone H2B has two flexible histone tails that extend outwards – one at the N-terminal end and one at C-terminal end. These are highly involved in condensing chromatin from the beads-on-a-string conformation to a 30-nm fiber. Similar to other histone proteins, histone H2B has a distinct histone fold that is optimized for histone-histone as well as histone-DNA interactions. Two copies of histone H2B come together with two copies each of histone H2A, histone H3, and histone ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H2A
Histone H2A is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells. The other histone proteins are: Histone H1, H1, Histone H2B, H2B, Histone H3, H3 and Histone H4, H4. Background Histones are proteins that package DNA into nucleosomes. Histones are responsible for maintaining the shape and structure of a nucleosome. One chromatin molecule is composed of at least one of each core histones per 100 base pairs of DNA. There are five families of histones known to date; these histones are termed H1/H5, H2A, H2B, H3, and H4. H2A is considered a core histone, along with H2B, H3 and H4. Core formation first occurs through the interaction of two H2A molecules. Then, H2A forms a Protein dimer, dimer with H2B; the core molecule is complete when H3-H4 also attaches to form a tetramer. Sequence variants Histone H2A is composed of non-allelic variants. The term "Histone H2A" is intentionally non-specific and refers to a variety of closely relat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H4
Histone H4 is one of the five main histone proteins involved in the structure of chromatin in eukaryote, eukaryotic cells. Featuring a main globular domain and a long N-terminus, N-terminal tail, H4 is involved with the structure of the nucleosome of the 'beads on a string' organization. Histone proteins are highly post-translationally modified. Covalently bonded modifications include acetylation and methylation of the N-terminal tails. These modifications may alter Gene expression, expression of genes located on DNA associated with its parent histone octamer. Histone H4 is an important protein in the structure and function of chromatin, where its sequence variants and variable modification states are thought to play a role in the dynamic and long term regulation of genes. Genetics Histone H4 is encoded in multiple genes at different loci including: HIST1H4A, HIST1H4B, HIST1H4C, HIST1H4D, HIST1H4E, HIST1H4F, HIST1H4G, HIST1H4H, HIST1H4I, HIST1H4J, HIST1H4K, HIST1H4L, HIST2H4A, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acids
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the Proteinogenic amino acid, 22 α-amino acids incorporated into proteins. Only these 22 appear in the genetic code of life. Amino acids can be classified according to the locations of the core structural functional groups (Alpha and beta carbon, alpha- , beta- , gamma- (γ-) amino acids, etc.); other categories relate to Chemical polarity, polarity, ionization, and side-chain group type (aliphatic, Open-chain compound, acyclic, aromatic, Chemical polarity, polar, etc.). In the form of proteins, amino-acid ''Residue (chemistry)#Biochemistry, residues'' form the second-largest component (water being the largest) of human muscles and other tissue (biology), tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-terminus
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, carboxy tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ... or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus. Chemistry Each amino acid has a carboxyl group and an amine group. Amino acids link to one another to form a chain by a dehydration reaction which joins the amine group of one amino acid to the carboxyl group of the next. Thus polypeptide chains have an end with an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |