|
HIF1α
Hypoxia-inducible factor 1-alpha, also known as HIF-1-alpha, is a subunit of a heterodimeric transcription factor hypoxia-inducible factor 1 (HIF-1) that is encoded by the ''HIF1A'' gene. The Nobel Prize in Physiology or Medicine 2019 was awarded for the discovery of HIF. HIF1A is a basic helix-loop-helix PAS domain containing protein, and is considered as the master transcriptional regulator of cellular and developmental response to hypoxia. The dysregulation and overexpression of ''HIF1A'' by either hypoxia or genetic alternations have been heavily implicated in cancer biology, as well as a number of other pathophysiologies, specifically in areas of vascularization and angiogenesis, energy metabolism, cell survival, and tumor invasion. The presence of HIF1A in a hypoxic environment is required to push forward normal placental development in early gestation. Two other alternative transcripts encoding different isoforms have been identified. Structure HIF1 is a heterod ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HIF-1
Hypoxia-inducible factors (HIFs) are transcription factors that respond to decreases in available oxygen in the cellular environment, or Hypoxia (medical), hypoxia. They also respond to instances of pseudohypoxia, such as thiamine deficiency. Both hypoxia and pseudohypoxia leads to impairment of adenosine triphosphate (ATP) production by the mitochondria. Discovery The HIF transcriptional complex was discovered in 1995 by Gregg L. Semenza and postdoctoral fellow Guang Wang. In 2016, William Kaelin Jr., Peter J. Ratcliffe and Gregg L. Semenza were presented the Lasker Award for their work in elucidating the role of HIF-1 in oxygen sensing and its role in surviving low oxygen conditions. In 2019, the same three individuals were jointly awarded the Nobel Prize in Physiology or Medicine for their work in elucidating how HIF senses and adapts cellular response to oxygen availability. Structure Oxygen-breathing species express the conservation (genetics), highly conserved tra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Subunit
In structural biology, a protein subunit is a polypeptide chain or single protein molecule that assembles (or "''coassembles''") with others to form a protein complex. Large assemblies of proteins such as viruses often use a small number of types of protein subunits as building blocks. A subunit is often named with a Greek or Roman letter, and the numbers of this type of subunit in a protein is indicated by a subscript. For example, ATP synthase has a type of subunit called α. Three of these are present in the ATP synthase molecule, leading to the designation α3. Larger groups of subunits can also be specified, like α3β3-hexamer and c-ring. Naturally occurring proteins that have a relatively small number of subunits are referred to as oligomeric.Quote: ''Oligomer molecule: A molecule of intermediate relative molecular mass, the structure of which essentially comprises a small plurality of units derived, actually or conceptually, from molecules of lower relative molecula ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isoforms
A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. (For that, see Proteoforms.) Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments (exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein. The discovery of isoforms could explain the discrepancy between the small number of protein coding regions of genes revealed by the human genome project and the large diversity of proteins seen in an organism: different proteins enc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Post-translational Modifications
In molecular biology, post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes, which translation (biology), translate mRNA into polypeptide chains, which may then change to form the mature protein product. PTMs are important components in cell signal transduction, signalling, as for example when prohormones are converted to hormones. Post-translational modifications can occur on the amino acid side chains or at the protein's C-terminus, C- or N-terminus, N- termini. They can expand the chemical set of the 22 proteinogenic amino acid, amino acids by changing an existing functional group or adding a new one such as phosphate. Phosphorylation is highly effective for controlling the enzyme activity and is the most common change after translation. Many eukaryotic and prokaryotic proteins also have carbohydrate molecules attached to them in a pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HIF Nobel Prize Physiology Medicine 2019 Hegasy ENG
HIF may refer to: Sports clubs * Habo IF, in Sweden * Hammarby IF, in Stockholm, Sweden * Hässleholms IF, in Sweden * Helsingborgs IF, in Sweden * Hemmingsmarks IF, in Sweden * Hörvikens IF, in Sölvesborg, Sweden * Hvidovre IF, in Denmark Other uses * Fiji Hindi (ISO 693-3 language code) * Finnmark University College (Norwegian: '), now part of the University of Tromsø * Harrogate International Festivals * Health Impact Fund, a proposal of incentives for global health * Health Insurance Fund, an Australian insurer * Hill Air Force Base, in Utah, United States * Horizontal Integration Facility * Hypoxia-inducible factor Hypoxia-inducible factors (HIFs) are transcription factors that respond to decreases in available oxygen in the cellular environment, or hypoxia. They also respond to instances of pseudohypoxia, such as thiamine deficiency. Both hypoxia an ... * USGS Hydrologic Instrumentation Facility, of the United States Geological Survey {{disambiguation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Redox
Redox ( , , reduction–oxidation or oxidation–reduction) is a type of chemical reaction in which the oxidation states of the reactants change. Oxidation is the loss of electrons or an increase in the oxidation state, while reduction is the gain of electrons or a decrease in the oxidation state. The oxidation and reduction processes occur simultaneously in the chemical reaction. There are two classes of redox reactions: * Electron transfer, Electron-transfer – Only one (usually) electron flows from the atom, ion, or molecule being oxidized to the atom, ion, or molecule that is reduced. This type of redox reaction is often discussed in terms of redox couples and electrode potentials. * Atom transfer – An atom transfers from one Substrate (chemistry), substrate to another. For example, in the rusting of iron, the oxidation state of iron atoms increases as the iron converts to an oxide, and simultaneously, the oxidation state of oxygen decreases as it accepts electrons r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GC-rich
In molecular biology and genetics, GC-content (or guanine-cytosine content) is the percentage of nitrogenous bases in a DNA or RNA molecule that are either guanine (G) or cytosine (C). This measure indicates the proportion of G and C bases out of an implied four total bases, also including adenine and thymine in DNA and adenine and uracil in RNA. GC-content may be given for a certain fragment of DNA or RNA or for an entire genome. When it refers to a fragment, it may denote the GC-content of an individual gene or section of a gene (domain), a group of genes or gene clusters, a non-coding region, or a synthetic oligonucleotide such as a primer. Structure Qualitatively, guanine (G) and cytosine (C) undergo a specific hydrogen bonding with each other, whereas adenine (A) bonds specifically with thymine (T) in DNA and with uracil (U) in RNA. Quantitatively, each GC base pair is held together by three hydrogen bonds, while AT and AU base pairs are held together by two hydrogen bonds. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Immunostaining
In biochemistry, immunostaining is any use of an antibody-based method to detect a specific protein in a sample. The term "immunostaining" was originally used to refer to the immunohistochemical staining of tissue sections, as first described by Albert Coons in 1941. However, immunostaining now encompasses a broad range of techniques used in histology, cell biology, and molecular biology that use antibody-based staining methods. Techniques Immunohistochemistry Immunohistochemistry or IHC staining of tissue sections (or immunocytochemistry, which is the staining of cells), is perhaps the most commonly applied immunostaining technique. While the first cases of IHC staining used fluorescent dyes (see ''immunofluorescence''), other non-fluorescent methods using enzymes such as peroxidase (see '' immunoperoxidase staining'') and alkaline phosphatase are now used. These enzymes are capable of catalysing reactions that give a coloured product that is easily detectable by ligh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
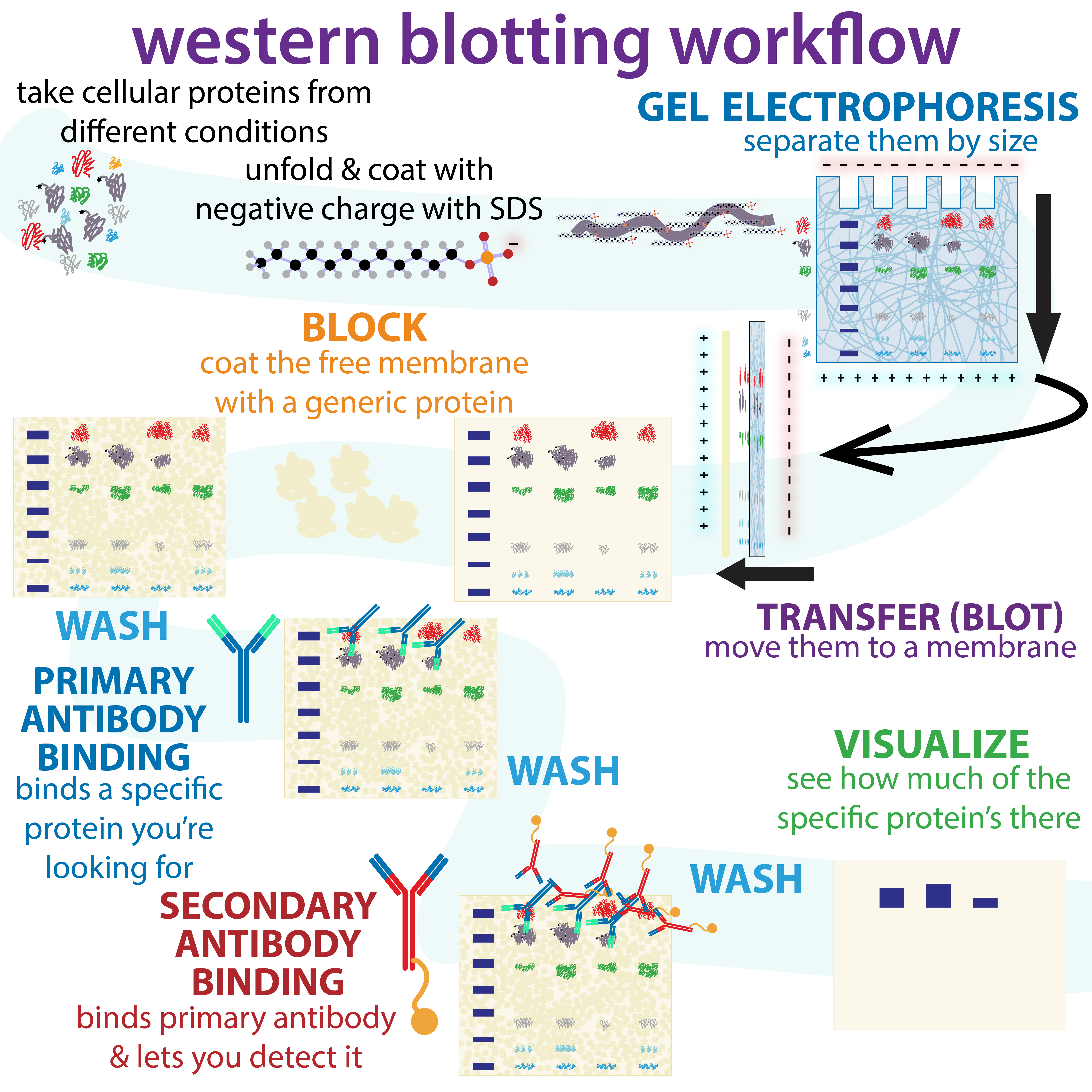
Western Blot
The western blot (sometimes called the protein immunoblot), or western blotting, is a widely used analytical technique in molecular biology and immunogenetics to detect specific proteins in a sample of tissue homogenate or extract. Besides detecting the proteins, this technique is also utilized to visualize, distinguish, and quantify the different proteins in a complicated protein combination. Western blot technique uses three elements to achieve its task of separating a specific protein from a complex: separation by size, transfer of protein to a solid support, and marking target protein using a primary and secondary antibody to visualize. A synthetic or animal-derived antibody (known as the primary antibody) is created that recognizes and binds to a specific target protein. The electrophoresis membrane is washed in a solution containing the primary antibody, before excess antibody is washed off. A secondary antibody is added which recognizes and binds to the primary antibod ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transactivating Domain
The transactivation domain or trans-activating domain (TAD) is a transcription factor scaffold domain which contains binding sites for other proteins such as transcription coregulators. These binding sites are frequently referred to as activation functions (AFs). TADs are named after their amino acid composition. These amino acids are either essential for the activity or simply the most abundant in the TAD. Transactivation by the Gal4 transcription factor is mediated by acidic amino acids, whereas hydrophobic residues in Gcn4 play a similar role. Hence, the TADs in Gal4 and Gcn4 are referred to as acidic or hydrophobic, respectively. In general we can distinguish four classes of TADs: * acidic domains (called also “acid blobs” or “negative noodles", rich in D and E amino acids, present in Gal4, Gcn4 and VP16). * glutamine-rich domains (contains multiple repetitions like "QQQXXXQQQ", present in SP1) * proline-rich domains (contains repetitions like "PPPXXXPPP" present in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Localization Signal
A nuclear localization signal ''or'' sequence (NLS) is an amino acid sequence that 'tags' a protein for import into the cell nucleus by nuclear transport. Typically, this signal consists of one or more short sequences of positively charged lysines or arginines exposed on the protein surface. Different nuclear localized proteins may share the same NLS. An NLS has the opposite function of a nuclear export signal (NES), which targets proteins out of the nucleus. Types Classical These types of NLSs can be further classified as either monopartite or bipartite. The major structural differences between the two are that the two basic amino acid clusters in bipartite NLSs are separated by a relatively short spacer sequence (hence bipartite - 2 parts), while monopartite NLSs are not. The first NLS to be discovered was the sequence PKKKRKV in the SV40 Large T-antigen (a monopartite NLS). The NLS of nucleoplasmin, KR AATKKAGQAKKK, is the prototype of the ubiquitous bipartite signal: two ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |