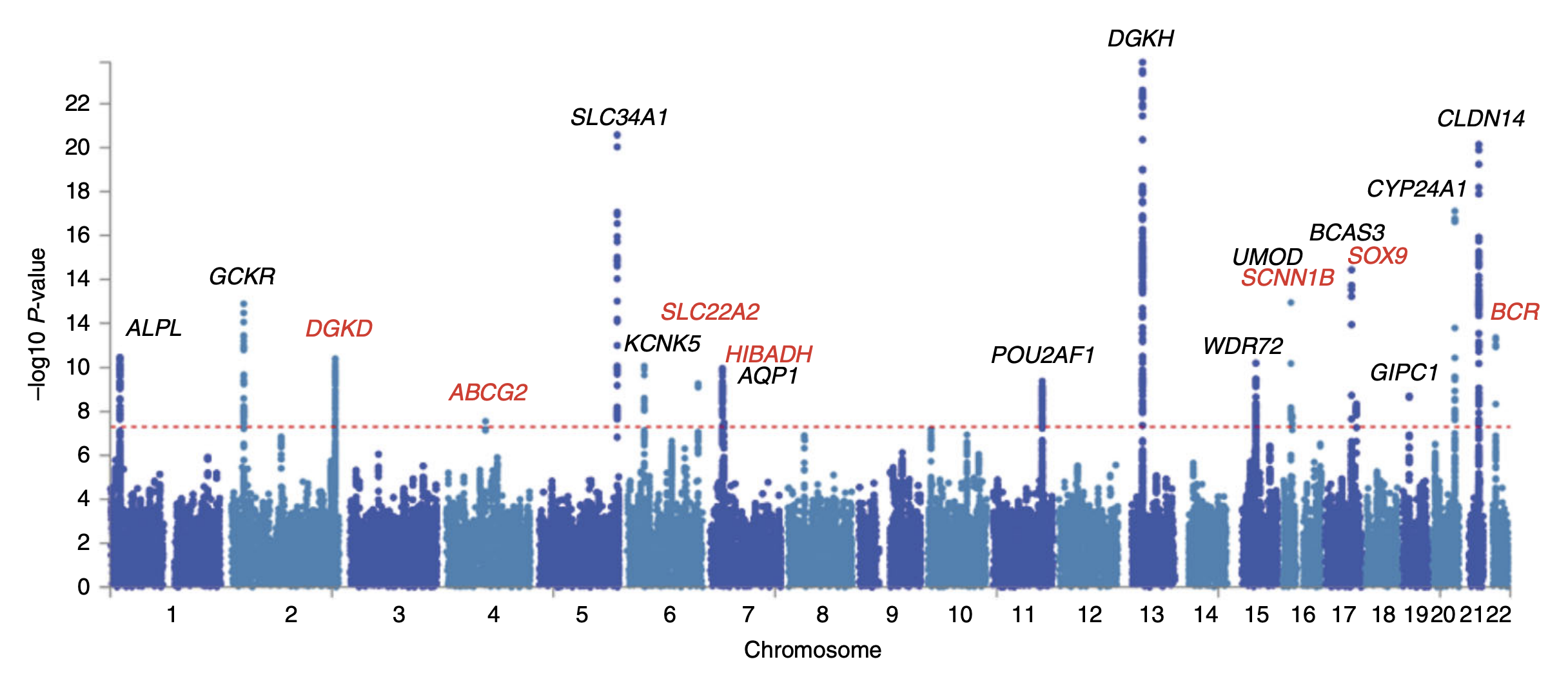
|
Genotyping By Sequencing
In the field of genetic sequencing, genotyping by sequencing, also called GBS, is a method to discover single nucleotide polymorphisms (SNP) in order to perform genotyping studies, such as genome-wide association studies (GWAS). GBS uses restriction enzymes to reduce genome complexity and genotype multiple DNA samples. After digestion, PCR is performed to increase fragments pool and then GBS libraries are sequenced using next generation sequencing technologies, usually resulting in about 100bp single-end reads. It is relatively inexpensive and has been used in plant breeding. Although GBS presents an approach similar to restriction-site-associated DNA sequencing (RAD-seq) method, they differ in some substantial ways. Methods GBS is a robust, simple, and affordable procedure for SNP discovery and mapping. Overall, this approach reduces genome complexity with restriction enzymes (REs) in high-diversity, large genomes species for efficient high-throughput, highly multiplexed sequen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid speed of sequencing attained with modern ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-nucleotide Polymorphism
In genetics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population (e.g. 1% or more), many publications do not apply such a frequency threshold. For example, at a specific base position in the human genome, the G nucleotide may appear in most individuals, but in a minority of individuals, the position is occupied by an A. This means that there is a SNP at this specific position, and the two possible nucleotide variations – G or A – are said to be the alleles for this specific position. SNPs pinpoint differences in our susceptibility to a wide range of diseases, for example age-related macular degeneration (a common SNP in the CFH gene is associated with increased risk of the disease) or nonalcoholic fatty liver disease (a SNP in the PNPLA3 gene is associated wit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genotyping
Genotyping is the process of determining differences in the genetic make-up (genotype) of an individual by examining the individual's DNA sequence using biological assays and comparing it to another individual's sequence or a reference sequence. It reveals the alleles an individual has inherited from their parents. Traditionally genotyping is the use of DNA sequences to define biological populations by use of molecular tools. It does not usually involve defining the genes of an individual. Techniques Current methods of genotyping include restriction fragment length polymorphism identification (RFLPI) of genomic DNA, random amplified polymorphic detection (RAPD) of genomic DNA, amplified fragment length polymorphism detection (AFLPD), polymerase chain reaction (PCR), DNA sequencing, allele specific oligonucleotide (ASO) probes, and hybridization to DNA microarrays or beads. Genotyping is important in research of genes and gene variants associated with disease. Due to curren ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome-wide Association Study
In genomics, a genome-wide association study (GWA study, or GWAS), also known as whole genome association study (WGA study, or WGAS), is an observational study of a genome-wide set of genetic variants in different individuals to see if any variant is associated with a trait. GWA studies typically focus on associations between single-nucleotide polymorphisms (SNPs) and traits like major human diseases, but can equally be applied to any other genetic variants and any other organisms. When applied to human data, GWA studies compare the DNA of participants having varying phenotypes for a particular trait or disease. These participants may be people with a disease (cases) and similar people without the disease (controls), or they may be people with different phenotypes for a particular trait, for example blood pressure. This approach is known as phenotype-first, in which the participants are classified first by their clinical manifestation(s), as opposed to genotype-first. Each pe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone (i.e. each strand) of the DNA double helix. These enzymes are found in bacteria and archaea and provide a defense mechanism against invading viruses. Inside a prokaryote, the restriction enzymes selectively cut up ''foreign'' DNA in a process called ''restriction digestion''; meanwhile, host DNA is protected by a modification enzyme (a methyltransferase) that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies (complete or partial) of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it (or a part of it) to a large enough amount to study in detail. PCR was invented in 1983 by the American biochemist Kary Mullis at Cetus Corporation; Mullis and biochemist Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications including biomedical resear ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plant Breeding
Plant breeding is the science of changing the traits of plants in order to produce desired characteristics. It has been used to improve the quality of nutrition in products for humans and animals. The goals of plant breeding are to produce crop varieties that boast unique and superior traits for a variety of applications. The most frequently addressed agricultural traits are those related to biotic and abiotic stress tolerance, grain or biomass yield, end-use quality characteristics such as taste or the concentrations of specific biological molecules (proteins, sugars, lipids, vitamins, fibers) and ease of processing (harvesting, milling, baking, malting, blending, etc.). Plant breeding can be performed through many different techniques ranging from simply selecting plants with desirable characteristics for propagation, to methods that make use of knowledge of genetics and chromosomes, to more complex molecular techniques. Genes in a plant are what determine what type of quali ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Site Associated DNA Markers
Restriction site associated DNA (RAD) markers are a type of genetic marker which are useful for association mapping, QTL-mapping, population genetics, ecological genetics and evolutionary genetics. The use of RAD markers for genetic mapping is often called RAD mapping. An important aspect of RAD markers and mapping is the process of isolating RAD tags, which are the DNA sequences that immediately flank each instance of a particular restriction site of a restriction enzyme throughout the genome. Once RAD tags have been isolated, they can be used to identify and genotype DNA sequence polymorphisms mainly in form of single nucleotide polymorphisms (SNPs). Polymorphisms that are identified and genotyped by isolating and analyzing RAD tags are referred to as RAD markers. Although genotyping by sequencing presents an approach similar to the RAD-seq method, they differ in some substantial ways. Isolation of RAD tags The use of the flanking DNA sequences around each restriction site is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Mass
The molecular mass (''m'') is the mass of a given molecule: it is measured in daltons (Da or u). Different molecules of the same compound may have different molecular masses because they contain different isotopes of an element. The related quantity relative molecular mass, as defined by IUPAC, is the ratio of the mass of a molecule to the unified atomic mass unit (also known as the dalton) and is unitless. The molecular mass and relative molecular mass are distinct from but related to the molar mass. The molar mass is defined as the mass of a given substance divided by the amount of a substance and is expressed in g/mol. That makes the molar mass an average of many particles or molecules, and the molecular mass the mass of one specific particle or molecule. The molar mass is usually the more appropriate figure when dealing with macroscopic (weigh-able) quantities of a substance. The definition of molecular weight is most authoritatively synonymous with relative molecular mass; ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sticky And Blunt Ends
DNA ends refer to the properties of the ends of linear DNA molecules, which in molecular biology are described as "sticky" or "blunt" based on the shape of the complementary strands at the terminus. In sticky ends, one strand is longer than the other (typically by at least a few nucleotides), such that the longer strand has bases which are left unpaired. In blunt ends, both strands are of equal length – i.e. they end at the same base position, leaving no unpaired bases on either strand. The concept is used in molecular biology, in cloning, or when subcloning insert DNA into vector DNA. Such ends may be generated by restriction enzymes that break the molecule's phosphodiester backbone at specific locations, which themselves belong to a larger class of enzymes called exonucleases and endonucleases. A restriction enzyme that cuts the backbones of both strands at non-adjacent locations leaves a staggered cut, generating two overlapping sticky ends, while an enzyme that makes a st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Burrows–Wheeler Aligner
This list of sequence alignment software is a compilation of software tools and web portals used in pairwise sequence alignment and multiple sequence alignment. See structural alignment software for structural alignment of proteins. Database search only *Sequence type: protein or nucleotide Pairwise alignment *Sequence type: protein or nucleotide **Alignment type: local or global Multiple sequence alignment *Sequence type: protein or nucleotide. **Alignment type: local or global Genomics analysis *Sequence type: protein or nucleotide Motif finding *Sequence type: protein or nucleotide Benchmarking Alignment viewers, editors Please see List of alignment visualization software. Short-read sequence alignment See also * List of open source bioinformatics software References {{Reflist Sequence Sequence alignment software This list of sequence alignment software is a compilation of software tools and web portals used in pairwise sequence alignment and multiple sequen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bowtie (sequence Analysis)
Bowtie is a software package commonly used for sequence alignment and sequence analysis in bioinformatics. The source code for the package is distributed freely and compiled binaries are available for Linux, macOS and Windows platforms. As of 2017, the ''Genome Biology'' paper describing the original Bowtie method has been cited more than 11,000 times. Bowtie is open-source software and is currently maintained by Johns Hopkins University. History The Bowtie sequence aligner was originally developed by Ben Langmead ''et al.'' at the University of Maryland in 2009. The aligner is typically used with short reads and a large reference genome, or for whole genome analysis. Bowtie is promoted as "an ultrafast, memory-efficient short aligner for short DNA sequences." The speed increase of Bowtie is partly due to implementing the Burrows–Wheeler transform for aligning, which reduces the memory footprint (typically to around 2.2GB for the human genome); a similar method is used by th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |