|
Exitron
Exitrons (exonic introns) are produced through alternative splicing and have characteristics of both introns and exons, but are described as retained introns. Even though they are considered introns, which are typically cut out of pre mRNA sequences, there are significant problems that arise when exitrons are spliced out of these strands, with the most obvious result being altered protein structures and functions. They were first discovered in plants, but have recently been found in metazoan species as well. Alternative splicing Exitrons are a result of alternative splicing (AS), in which introns are typically cut out of a pre mRNA sequence, while exons remain in the sequence and are translated into proteins. The same sequence within a pre mRNA strand can be considered an intron or exon depending on the desired protein to be produced. As a result, different final mRNA sequences are generated and a large variety of proteins can be made from one single gene. Mutations that exist in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome. History The term ''exon'' is a shortening of the phrase ''expressed region'' and was coined by American biochemist Walter Gilbert in 1978: "The notion of the cistron... must be replaced by that of a transcription unit containing regions which will be lost from the mature messengerwhich I suggest we call introns (for intragenic regions)alternating with regions which will be expressedexons." This definition was originally made for protein-coding transcripts that are spliced before b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of the cistron .e., gene... must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger – which I suggest we call introns (for intragenic regions) – alternating with regions which will be expressed – exons." (Gilbert 1978) The term ''intron'' refers to both the DNA sequence within a gene and the corresponding RNA sequence in RNA transcripts. The non-intron sequences that become joined by this RNA processing to form the mature RNA are called exons. Introns are found in the genes of most eukaryotes and many eukaryotic viruses, and they can be located in both protein-coding genes and genes that function as RNA ( noncoding genes). There are four main types of introns: tRNA introns, group I introns, group I ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alternative Splicing
Alternative splicing, alternative RNA splicing, or differential splicing, is an alternative RNA splicing, splicing process during gene expression that allows a single gene to produce different splice variants. For example, some exons of a gene may be included within or excluded from the final RNA product of the gene. This means the exons are joined in different combinations, leading to different splice variants. In the case of protein-coding genes, the proteins translated from these splice variants may contain differences in their amino acid sequence and in their biological functions (see Figure). Biologically relevant alternative splicing occurs as a normal phenomenon in eukaryotes, where it increases the number of proteins that can be encoded by the genome. In humans, it is widely believed that ~95% of multi-exonic genes are alternatively spliced to produce functional alternative products from the same gene but many scientists believe that most of the observed splice variants ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
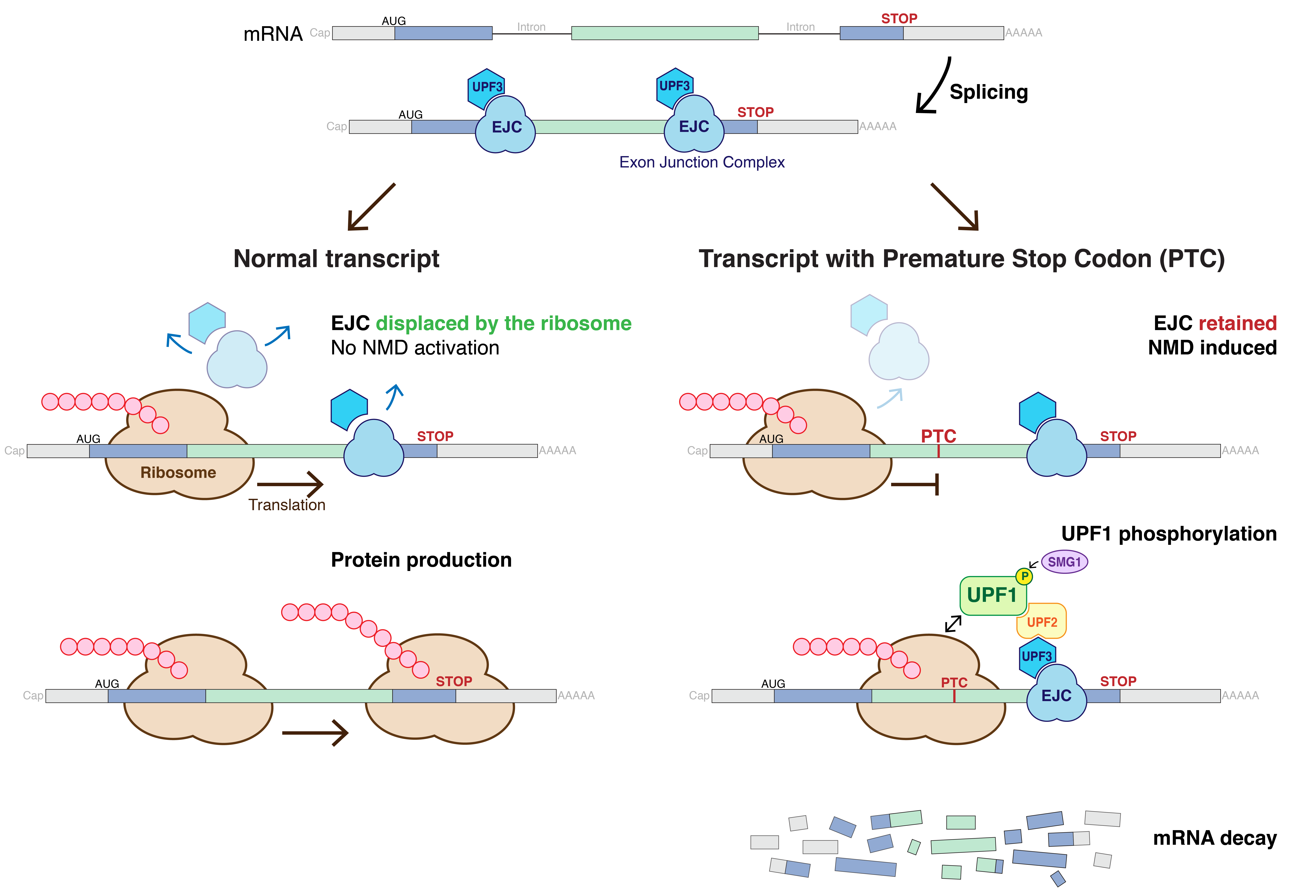
Nonsense-mediated Decay
Nonsense-mediated mRNA decay (NMD) is a surveillance pathway that exists in all eukaryotes. Its main function is to reduce errors in gene expression by eliminating mRNA transcripts that contain premature stop codons. Translation of these aberrant mRNAs could, in some cases, lead to deleterious gain-of-function or dominant-negative activity of the resulting proteins. NMD was first described in human cells and in yeast almost simultaneously in 1979. This suggested broad phylogenetic conservation and an important biological role of this intriguing mechanism. NMD was discovered when it was realized that cells often contain unexpectedly low concentrations of mRNAs that are transcribed from alleles carrying nonsense mutations. Nonsense mutations code for a premature stop codon which causes the protein to be shortened. The truncated protein may or may not be functional, depending on the severity of what is not translated. In human genetics, NMD has the possibility to not only limit t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteome Assortment
A proteome is the entire set of proteins that is, or can be, expressed by a genome, cell, tissue, or organism at a certain time. It is the set of expressed proteins in a given type of cell or organism, at a given time, under defined conditions. Proteomics is the study of the proteome. Types of proteomes While proteome generally refers to the proteome of an organism, multicellular organisms may have very different proteomes in different cells, hence it is important to distinguish proteomes in cells and organisms. A cellular proteome is the collection of proteins found in a particular cell type under a particular set of environmental conditions such as exposure to hormone stimulation. It can also be useful to consider an organism's complete proteome, which can be conceptualized as the complete set of proteins from all of the various cellular proteomes. This is very roughly the protein equivalent of the genome. The term ''proteome'' has also been used to refer to the collection o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Generalized Additive Model
In statistics, a generalized additive model (GAM) is a generalized linear model in which the linear response variable depends linearly on unknown smooth functions of some predictor variables, and interest focuses on inference about these smooth functions. GAMs were originally developed by Trevor Hastie and Robert Tibshirani to blend properties of generalized linear models with additive models. They can be interpreted as the discriminative generalization of the naive Bayes generative model. The model relates a univariate response variable, ''Y'', to some predictor variables, ''x''''i''. An exponential family distribution is specified for Y (for example normal, binomial or Poisson distributions) along with a link function ''g'' (for example the identity or log functions) relating the expected value of ''Y'' to the predictor variables via a structure such as : g(\operatorname(Y))=\beta_0 + f_1(x_1) + f_2(x_2)+ \cdots + f_m(x_m).\,\! The functions ''f''''i'' may be functions with ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
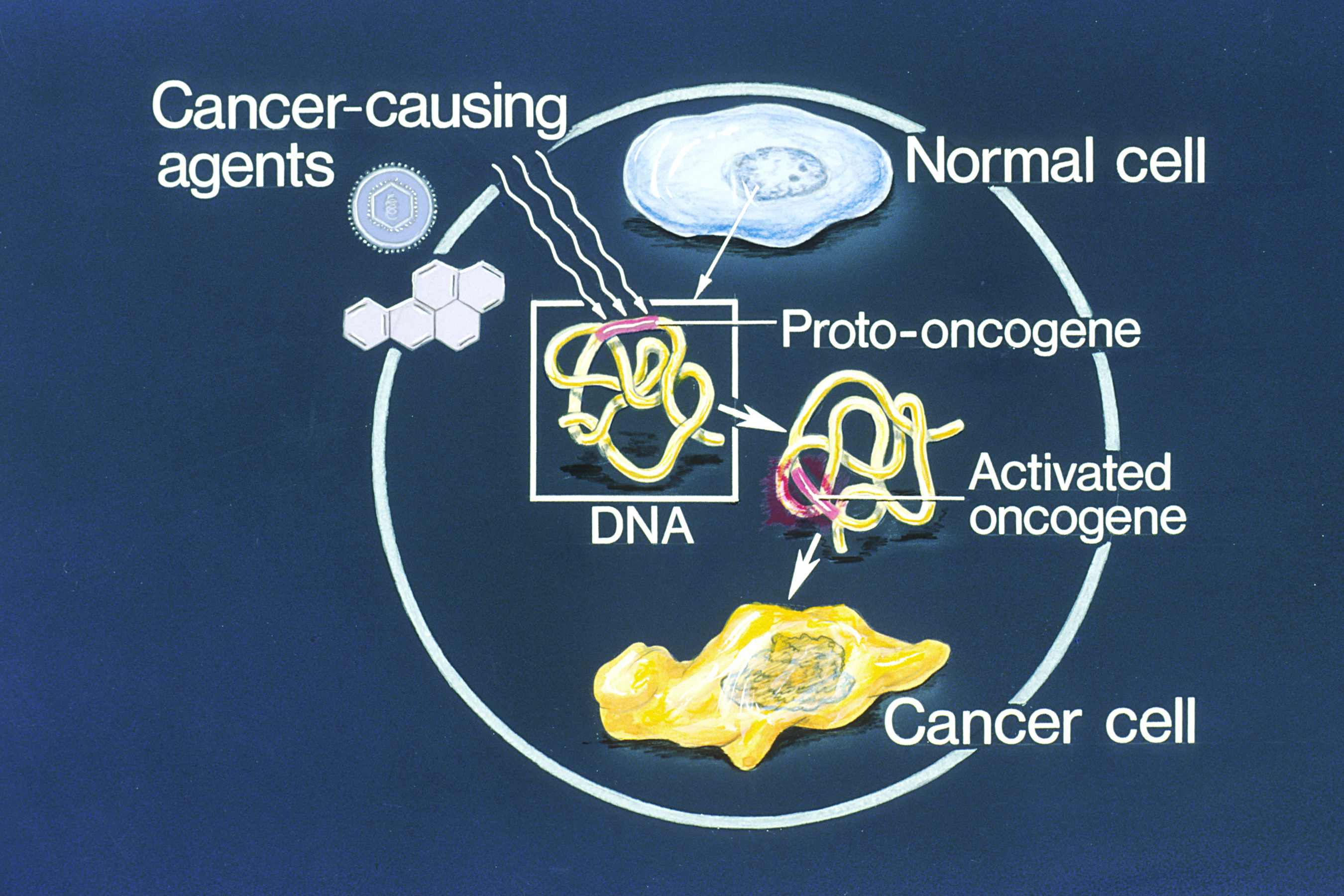
Oncogenes
An oncogene is a gene that has the potential to cause cancer. In tumor cells, these genes are often mutated, or expressed at high levels.Kimball's Biology Pages. "Oncogenes" Free full text Most normal cells undergo a preprogrammed rapid cell death () if critical functions are altered and then malfunction. Activated oncogenes can cause those cells designated for apoptosis to survive and proliferate instead. Most oncogenes began as proto-oncogenes: normal genes involved in cell growth and proliferation or inhibition of apoptosis. If, through mutation, normal genes promoting cellular growth are up-regulated (gain-of-function mutation), they predispose the cel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
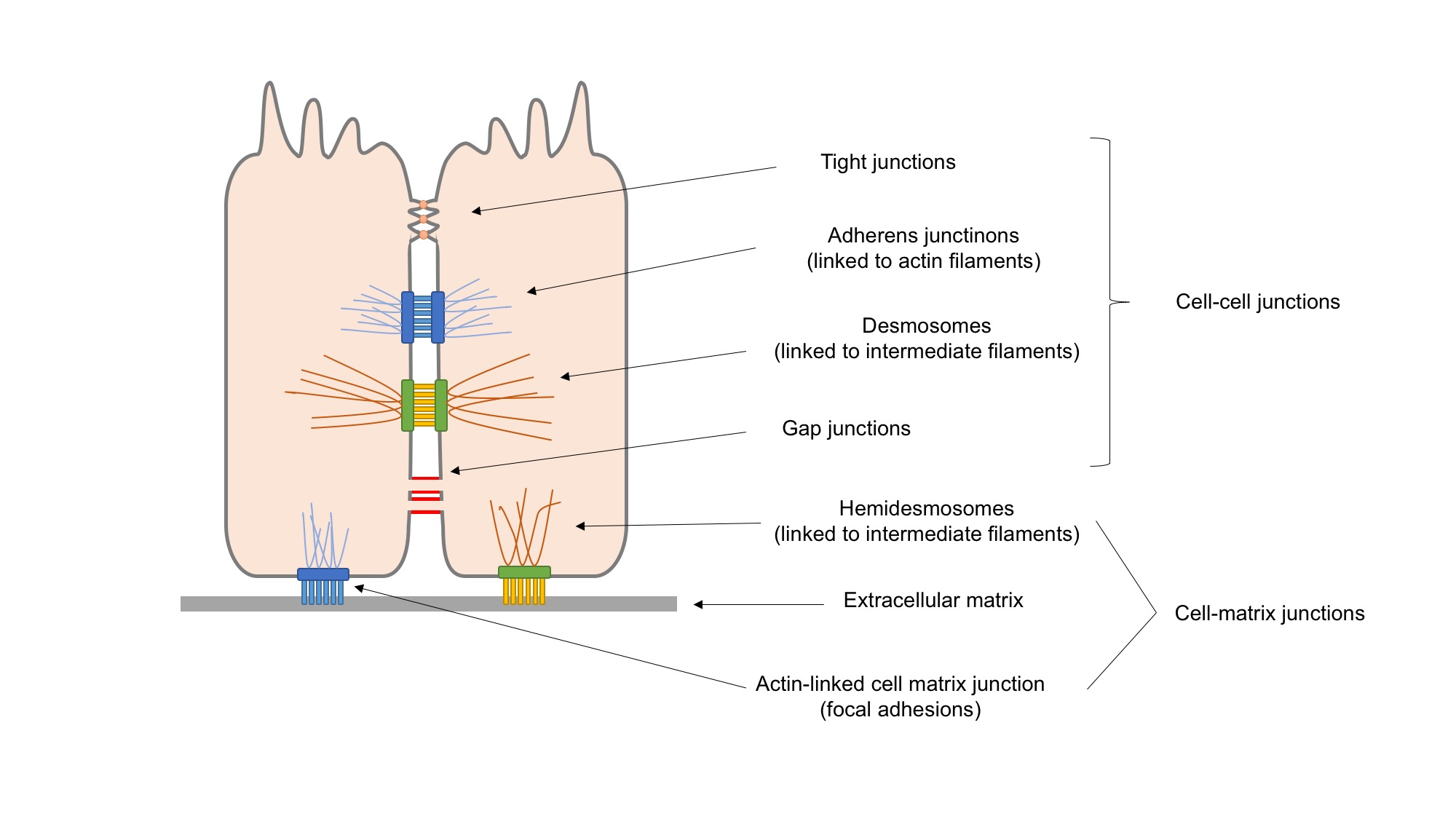
Cell Adhesion
Cell adhesion is the process by which cells interact and attach to neighbouring cells through specialised molecules of the cell surface. This process can occur either through direct contact between cell surfaces such as Cell_junction, cell junctions or indirect interaction, where cells attach to surrounding extracellular matrix (ECM), a gel-like structure containing molecules released by cells into spaces between them. Cells adhesion occurs from the interactions between cell adhesion molecules, cell-adhesion molecules (CAMs), transmembrane proteins located on the cell surface. Cell adhesion links cells in different ways and can be involved in signal transduction for cells to detect and respond to changes in the surroundings. Other cellular processes regulated by cell adhesion include cell migration and tissue development in multicellular organisms. Alterations in cell adhesion can disrupt important cellular processes and lead to a variety of diseases, including cancer and arthrit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell Migration
Cell migration is a central process in the development and maintenance of multicellular organisms. Tissue formation during embryogenesis, embryonic development, wound healing and immune system, immune responses all require the orchestrated movement of cells in particular directions to specific locations. Cells often migrate in response to specific external signals, including chemotaxis, chemical signals and mechanotaxis, mechanical signals. Errors during this process have serious consequences, including intellectual disability, cardiovascular disease, vascular disease, tumor, tumor formation and metastasis. An understanding of the mechanism by which cells migrate may lead to the development of novel therapeutic strategies for controlling, for example, invasive tumour cells. Due to the highly viscous environment (low Reynolds number), cells need to continuously produce forces in order to move. Cells achieve active movement by very different mechanisms. Many less complex prokaryotic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metastasis
Metastasis is a pathogenic agent's spreading from an initial or primary site to a different or secondary site within the host's body; the term is typically used when referring to metastasis by a cancerous tumor. The newly pathological sites, then, are metastases (mets). It is generally distinguished from cancer invasion, which is the direct extension and penetration by cancer cells into neighboring tissues. Cancer occurs after cells are genetically altered to proliferate rapidly and indefinitely. This uncontrolled proliferation by mitosis produces a primary tumor, primary tumour heterogeneity, heterogeneic tumour. The cells which constitute the tumor eventually undergo metaplasia, followed by dysplasia then anaplasia, resulting in a Malignancy, malignant phenotype. This malignancy allows for invasion into the circulation, followed by invasion to a second site for tumorigenesis. Some cancer cells, known as circulating tumor cells (CTCs), are able to penetrate the walls of lymp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Spliceosome
A spliceosome is a large ribonucleoprotein (RNP) complex found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs ( snRNA) and numerous proteins. Small nuclear RNA (snRNA) molecules bind to specific proteins to form a small nuclear ribonucleoprotein complex (snRNP, pronounced "snurps"), which in turn combines with other snRNPs to form a large ribonucleoprotein complex called a spliceosome. The spliceosome removes introns from a transcribed pre-mRNA, a type of primary transcript. This process is generally referred to as splicing. An analogy is a film editor, who selectively cuts out irrelevant or incorrect material (equivalent to the introns) from the initial film and sends the cleaned-up version to the director for the final cut. However, sometimes the RNA within the intron acts as a ribozyme, splicing itself without the use of a spliceosome or protein enzymes. History In 1977, work by the Sharp and Roberts labs reve ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |