|
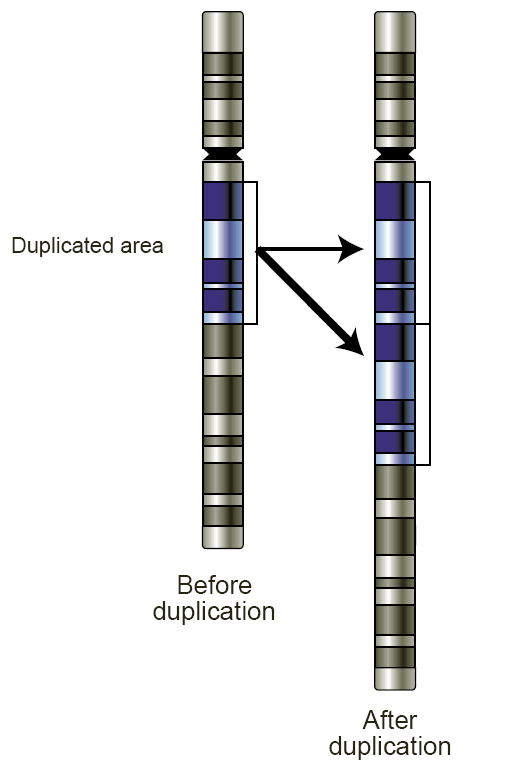
Copy Number Variation
Copy number variation (CNV) is a phenomenon in which sections of the genome are repeated and the number of repeats in the genome varies between individuals. Copy number variation is a type of structural variation: specifically, it is a type of duplication or deletion event that affects a considerable number of base pairs. Approximately two-thirds of the entire human genome may be composed of repeats and 4.8–9.5% of the human genome can be classified as copy number variations. In mammals, copy number variations play an important role in generating necessary variation in the population as well as disease phenotype. Copy number variations can be generally categorized into two main groups: short repeats and long repeats. However, there are no clear boundaries between the two groups and the classification depends on the nature of the loci of interest. Short repeats include mainly dinucleotide repeats (two repeating nucleotides e.g. A-C-A-C-A-C...) and trinucleotide repeats. Lon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Expression
Gene expression is the process (including its Regulation of gene expression, regulation) by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, and ultimately affect a phenotype. These products are often proteins, but in non-protein-coding genes such as Transfer RNA, transfer RNA (tRNA) and Small nuclear RNA, small nuclear RNA (snRNA), the product is a functional List of RNAs, non-coding RNA. The process of gene expression is used by all known life—eukaryotes (including multicellular organisms), prokaryotes (bacteria and archaea), and viruses—to generate the macromolecule, macromolecular machinery for life. In genetics, gene expression is the most fundamental level at which the genotype gives rise to the phenotype, ''i.e.'' observable trait. The genetic information stored in DNA represents the genotype, whereas the phenotype results from the "interpretation" of that informati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role in the expression of genes. Because it is tightly packed, it was thought to be inaccessible to polymerases and therefore not transcribed; however, according to Volpe et al. (2002), and many other papers since, much of this DNA is in fact transcribed, but it is continuously turned over via RNA-induced transcriptional silencing (RITS). Recent studies with electron microscopy and OsO4 staining reveal that the dense packing is not due to the chromatin. Constitutive heterochromatin can affect the genes near itself (e.g. position-effect variegation). It is usually repetitive and forms structural functions such as centromeres or telomeres, in addition to acting as an attractor for other gene-expression or repression signals. Facultativ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Centromere
The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fibers attach to the centromere via the kinetochore. The physical role of the centromere is to act as the site of assembly of the kinetochores – a highly complex multiprotein structure that is responsible for the actual events of chromosome segregation – i.e. binding microtubules and signaling to the cell cycle machinery when all chromosomes have adopted correct attachments to the spindle, so that it is safe for cell division to proceed to completion and for cells to enter anaphase. There are, broadly speaking, two types of centromeres. "Point centromeres" bind to specific proteins that recognize particular DNA sequences with high efficiency. Any piece of DNA with the point centromere DNA sequence on it will typically form a centr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Telomere
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes (see #Sequences, Sequences). Telomeres are a widespread genetic feature most commonly found in eukaryotes. In most, if not all species possessing them, they protect the terminal regions of DNA, chromosomal DNA from progressive degradation and ensure the integrity of linear chromosomes by preventing DNA repair systems from mistaking the very ends of the DNA strand for a double-strand break. Discovery The existence of a special structure at the ends of chromosomes was independently proposed in 1938 by Hermann Joseph Muller, studying the fruit fly ''Drosophila melanogaster'', and in 1939 by Barbara McClintock, working with maize. Muller observed that the ends of irradiated fruit fly chromosomes did not present alterations such as deletions or inversions. He hypothesized the presence of a protective cap, which he coined "telomeres", from the Greek ' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microsatellite
A microsatellite is a tract of repetitive DNA in which certain Sequence motif, DNA motifs (ranging in length from one to six or more base pairs) are repeated, typically 5–50 times. Microsatellites occur at thousands of locations within an organism's genome. They have a higher mutation rate than other areas of DNA leading to high genetic diversity. Microsatellites are often referred to as short tandem repeats (STRs) by forensic genetics, forensic geneticists and in genetic genealogy, or as simple sequence repeats (SSRs) by plant geneticists. Microsatellites and their longer cousins, the minisatellites, together are classified as variable number tandem repeat, VNTR (variable number of tandem repeats) DNA. The name Satellite DNA, "satellite" DNA refers to the early observation that centrifugation of genomic DNA in a test tube separates a prominent layer of bulk DNA from accompanying "satellite" layers of repetitive DNA. They are widely used for DNA profiling in Loss of heterozygos ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
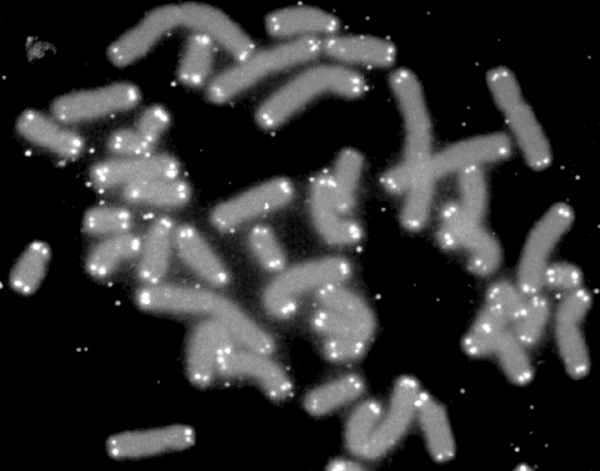
Fluorescent In Situ Hybridization
Fluorescence ''in situ'' hybridization (FISH) is a cytogenetics, molecular cytogenetic technique that uses hybridization probe, fluorescent probes that bind to only particular parts of a nucleic acid sequence with a high degree of sequence Complementarity (molecular biology), complementarity. It was developed by biomedical researchers in the early 1980s to detect and localize the presence or absence of specific DNA DNA sequence, sequences on chromosomes. Fluorescence microscopy can be used to find out where the fluorescent probe is bound to the chromosomes. FISH is often used for finding specific features in DNA for use in genetic counseling, medicine, and species identification. FISH can also be used to detect and localize specific RNA targets (mRNA, Long non-coding RNA, lncRNA and miRNA) in cells, circulating tumor cells, and tissue samples. In this context, it can help define the spatial-temporal patterns of gene expression within cells and tissues. Probes – RNA and DNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Diseases
A genetic disorder is a health problem caused by one or more abnormalities in the genome. It can be caused by a mutation in a single gene (monogenic) or multiple genes (polygenic) or by a chromosome abnormality. Although polygenic disorders are the most common, the term is mostly used when discussing disorders with a single genetic cause, either in a gene or chromosome. The mutation responsible can occur spontaneously before embryonic development (a ''de novo'' mutation), or it can be inherited from two parents who are carriers of a faulty gene (autosomal recessive inheritance) or from a parent with the disorder (autosomal dominant inheritance). When the genetic disorder is inherited from one or both parents, it is also classified as a hereditary disease. Some disorders are caused by a mutation on the X chromosome and have X-linked inheritance. Very few disorders are inherited on the Y chromosome or mitochondrial DNA (due to their size). There are well over 6,000 known geneti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Variation
Genetic variation is the difference in DNA among individuals or the differences between populations among the same species. The multiple sources of genetic variation include mutation and genetic recombination. Mutations are the ultimate sources of genetic variation, but other mechanisms, such as genetic drift, contribute to it, as well. Among individuals within a population Genetic variation can be identified at many levels. Identifying genetic variation is possible from observations of phenotypic variation in either quantitative traits (traits that vary continuously and are coded for by many genes, e.g., leg length in dogs) or discrete traits (traits that fall into discrete categories and are coded for by one or a few genes, e.g., white, pink, or red petal color in certain flowers). Genetic variation can also be identified by examining variation at the level of enzymes using the process of protein electrophoresis. Polymorphic genes have more than one allele at each locu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromosomal Rearrangement
In genetics, a chromosomal rearrangement is a mutation that is a type of chromosome abnormality involving a change in the structure of the native chromosome. Such changes may involve several different classes of events, like deletions, duplications, inversions, and translocations. Usually, these events are caused by a breakage in the DNA double helices at two different locations, followed by a rejoining of the broken ends to produce a new chromosomal arrangement of genes, different from the gene order of the chromosomes before they were broken. Structural chromosomal abnormalities are estimated to occur in around 0.5% of newborn infants. Some chromosomal regions are more prone to rearrangement than others and thus are the source of genetic diseases and cancer. This instability is usually due to the propensity of these regions to misalign during DNA repair, exacerbated by defects of the appearance of replication proteins (like FEN1 or Pol δ) that ubiquitously affect the inte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tandem
Tandem, or in tandem, is an arrangement in which two or more animals, machines, or people are lined up one behind another, all facing in the same direction. ''Tandem'' can also be used more generally to refer to any group of persons or objects working together, not necessarily in line. The English word ''tandem'' derives from the Latin adverb , meaning ''at length'' or ''finally''. It is a word play, using the Latin phrase (referring to time, not position) for English "at length, lengthwise". Horse driving When Driving (horse), driving horses, ''tandem'' refers to one horse harnessed in front of another to pull a load or Horse-drawn vehicle, vehicle. A tandem arrangement provides more pulling power than a single horse, such as for pulling a heavy load up a steep hill, out of heavy mud or snow, or pulling heavy loads on narrow tracks or through narrow gates and doorways (too wide for a pair of horses side-by-side). For example, a Brewer's van fully loaded with 25 barrels migh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Segmental Duplication
Low copy repeats (LCRs), also known as segmental duplications (SDs), or duplicons, are DNA sequences present in multiple locations within a genome that share high levels of sequence identity. Repeats The repeats, or duplications, are typically 10–300 kb in length, and bear greater than 95% sequence identity. Though rare in most mammals, LCRs comprise a large portion of the human genome owing to a significant expansion during primate evolution. In humans, chromosomes Y and 22 have the greatest proportion of SDs: 50.4% and 11.9% respectively. SRGAP2 is an SD. Misalignment of LCRs during non-allelic homologous recombination (NAHR) is an important mechanism underlying the chromosomal microdeletion disorders as well as their reciprocal duplication partners. Many LCRs are concentrated in "hotspots", such as the 17p11-12 region, 27% of which is composed of LCR sequence. NAHR and non-homologous end joining (NHEJ) within this region are responsible for a wide range of disorders, in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |