|
CTLA4
Cytotoxic T-lymphocyte associated protein 4, (CTLA-4) also known as CD152 (cluster of differentiation 152), is a protein receptor that functions as an immune checkpoint and downregulates immune responses. CTLA-4 is constitutively expressed in regulatory T cells but only upregulated in conventional T cells after activation – a phenomenon which is particularly notable in cancers. It acts as an "off" switch when bound to CD80 or CD86 on the surface of antigen-presenting cells. It is encoded by the gene ''CTLA4'' in humans. The CTLA-4 protein is encoded by the ''Ctla-4'' gene in mice. History CTLA-4 was first identified in 1991 as a second receptor for the T cell costimulation ligand B7. In November 1995, the labs of Tak Wah Mak and Arlene Sharpe independently published their findings on the discovery of the function of CTLA-4 as a negative regulator of T-cell activation, by knocking out the gene in mice. Previous studies from several labs had used methods which could not ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CD28
CD28 (Cluster of Differentiation 28) is a protein expressed on T cells that provides essential co-stimulation, co-stimulatory signals required for T cell activation and survival. When T cells are stimulated through CD28 in conjunction with the T-cell receptor (T cell receptor, TCR), it enhances the production of various interleukins, particularly interleukin 6, IL-6. CD28 serves as a receptor for CD80 (B7.1) and CD86 (B7.2), proteins found on antigen-presenting cells (APCs). CD28 is the only B7 (protein), B7 receptor consistently expressed on naive T cells. In the absence of CD28:B7 interaction, a naive T cell's TCR engagement with an Major histocompatibility complex, MHC:antigen complex leads to anergy. CD28 is also expressed on bone marrow stromal cells, plasma cells, neutrophils, and eosinophils, although its function in these cells is not fully understood. Typically, CD28 is expressed on about 50% of CD8, CD8+ T cells and more than 80% of CD4, CD4+ T cells in humans. However, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Immune Checkpoint
Immune checkpoints are regulators of the immune system. These pathways are crucial for self-tolerance, which prevents the immune system from attacking cells indiscriminately. However, some cancers can protect themselves from attack by stimulating immune checkpoint targets. Inhibitory checkpoint molecules are targets for cancer immunotherapy due to their potential for use in multiple types of cancers. Currently approved checkpoint inhibitors block CTLA4, PD-1 and PD-L1. For the related basic science discoveries, James P. Allison and Tasuku Honjo won the Tang Prize, Tang Prize in Biopharmaceutical Science and the Nobel Prize in Physiology or Medicine in 2018. Stimulatory checkpoint molecules Four stimulatory checkpoint molecules are members of the TNF receptor superfamily, tumor necrosis factor (TNF) receptor superfamily—CD27, CD40, OX40, GITR and CD137. Another two stimulatory checkpoint molecules belong to the B7-CD28 superfamily—CD28 itself and ICOS. * CD27: This molec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CD86
Cluster of Differentiation 86 (also known as CD86 and B7-2) is a protein constitutively expressed on dendritic cells, Langerhans cells, macrophages, B-cells (including memory B-cells), and on other antigen-presenting cells. Along with CD80, CD86 provides costimulatory signals necessary for T cell activation and survival. Depending on the ligand bound, CD86 can signal for self-regulation and cell-cell association, or for attenuation of regulation and cell-cell disassociation. The ''CD86'' gene encodes a type I membrane protein that is a member of the immunoglobulin superfamily. Alternative splicing results in two transcript variants encoding different isoforms. Additional transcript variants have been described, but their full-length sequences have not been determined. Structure CD86 belongs to the B7 family of the immunoglobulin superfamily. It is a 70 kDa glycoprotein made up of 329 amino acids. Both CD80 and CD86 share a conserved amino acid motif that forms their l ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Regulatory T Cell
The regulatory T cells (Tregs or Treg cells), formerly known as suppressor T cells, are a subpopulation of T cells that modulate the immune system, maintain immune tolerance, tolerance to self-antigens, and prevent autoimmune disease. Treg cells are immunosuppression, immunosuppressive and generally suppress or downregulation and upregulation, downregulate induction and proliferation of effector T cells. Treg cells express the biomarkers CD4, FOXP3, and CD25 and are thought to be derived from the same cell lineage, lineage as naïve T helper cell, CD4+ cells. Because effector T cells also express CD4 and CD25, Treg cells are very difficult to effectively discern from effector CD4+, making them difficult to study. Research has found that the cytokine Transforming growth factor beta, transforming growth factor beta (TGF-β) is essential for Treg cells to differentiate from naïve CD4+ cells and is important in maintaining Treg cell homeostas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Immunoglobulin V-set Domain
V-set domains are Ig-like domains resembling the antibody variable domain. V-set domains are found in diverse protein families, including immunoglobulin light and heavy chains; in several T-cell receptors such as CD2, CD4, CD80, and CD86; in the N-terminal part of Siglec-receptors, in myelin membrane adhesion molecules; in junctional adhesion molecules (JAM); in tyrosine-protein kinase receptors; and in the programmed cell death protein 1 (PD1). Subfamilies * Immunoglobulin V-set, subgroup * T-cell surface antigen CD2 Human proteins containing this domain ACAM; ACAN; ADAMTSL1; AGC1; AMICA1; BCAM; BCAN; BGP; BGPc; BT3.3; BTN1A1; BTN2A1; BTN2A2; BTN2A3; BTN3A1; BTN3A2; BTN3A3; BTNL2; BTNL3; BTNL8; BTNL9; C10orf54; C1orf32; C9orf94; CADM1; CADM2; CADM3; CADM4; CD2; CD226; CD274; CD276; CD300A; CD300C; CD300D; CD300E; CD300LB; ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
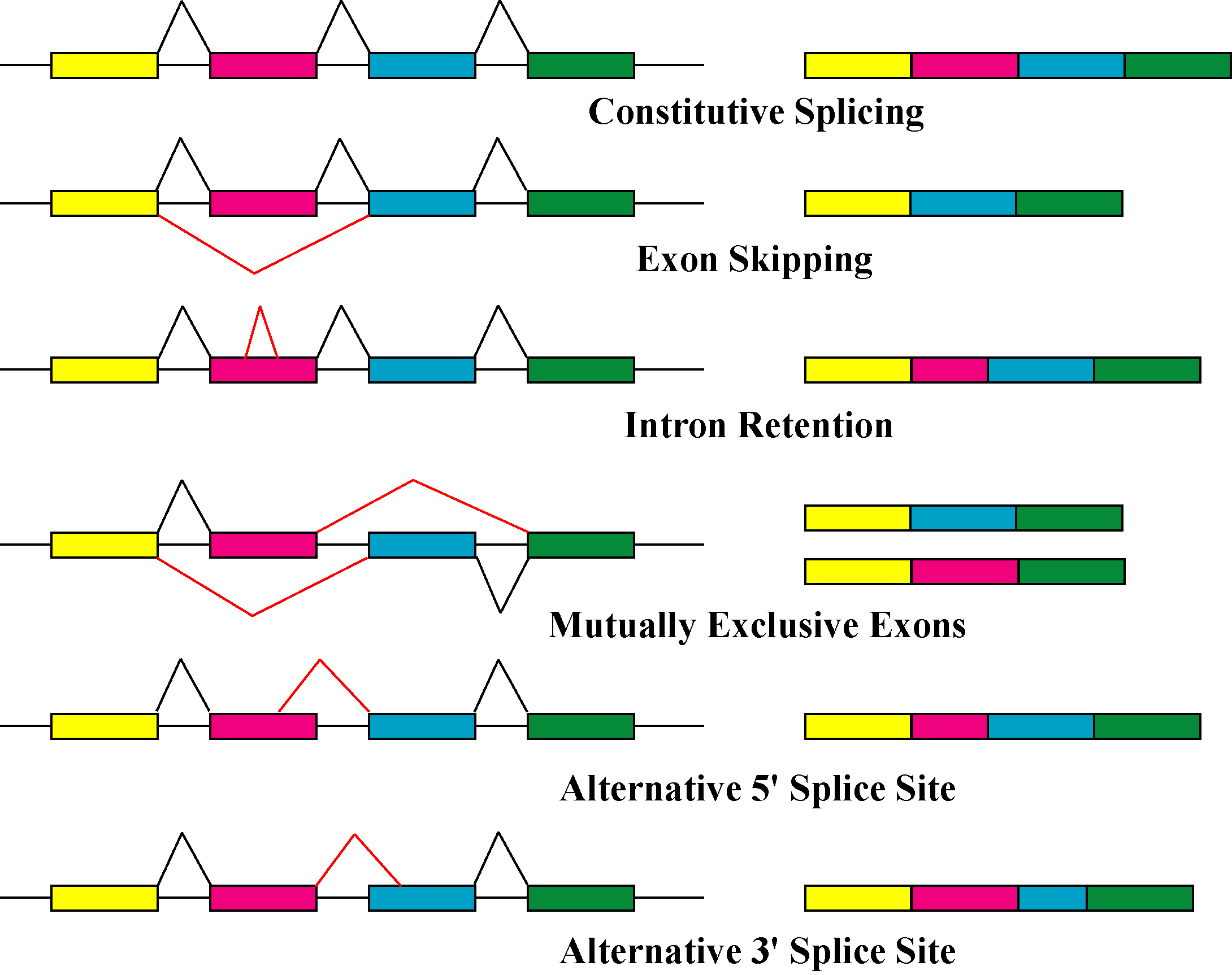
Alternative Splicing
Alternative splicing, alternative RNA splicing, or differential splicing, is an alternative RNA splicing, splicing process during gene expression that allows a single gene to produce different splice variants. For example, some exons of a gene may be included within or excluded from the final RNA product of the gene. This means the exons are joined in different combinations, leading to different splice variants. In the case of protein-coding genes, the proteins translated from these splice variants may contain differences in their amino acid sequence and in their biological functions (see Figure). Biologically relevant alternative splicing occurs as a normal phenomenon in eukaryotes, where it increases the number of proteins that can be encoded by the genome. In humans, it is widely believed that ~95% of multi-exonic genes are alternatively spliced to produce functional alternative products from the same gene but many scientists believe that most of the observed splice variants ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Extracellular
This glossary of biology terms is a list of definitions of fundamental terms and concepts used in biology, the study of life and of living organisms. It is intended as introductory material for novices; for more specific and technical definitions from sub-disciplines and related fields, see Glossary of cell biology, Glossary of genetics, Glossary of evolutionary biology, Glossary of ecology, Glossary of environmental science and Glossary of scientific naming, or any of the organism-specific glossaries in :Glossaries of biology. A B C D E ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
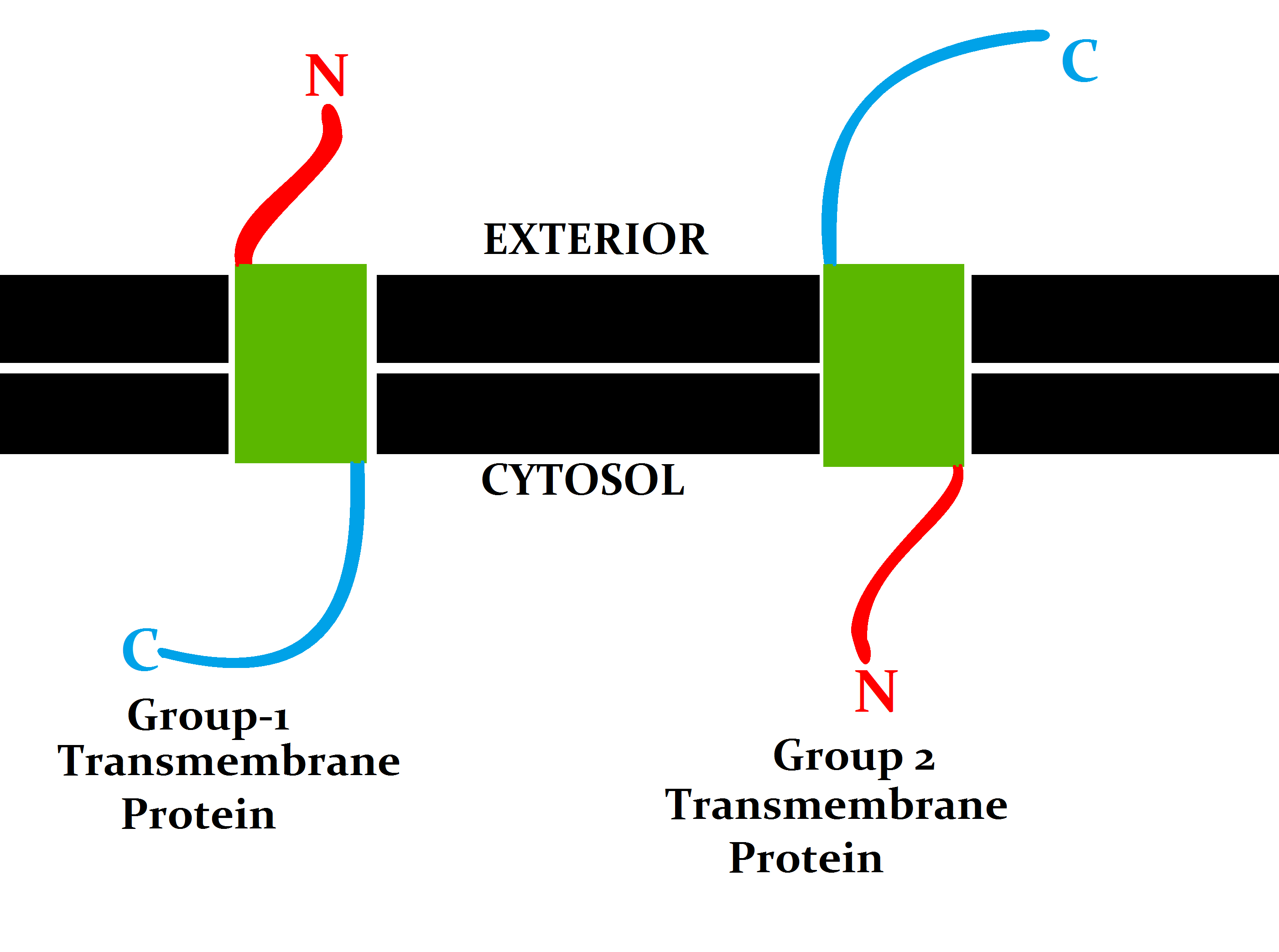
Transmembrane Protein
A transmembrane protein is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them ( beta-barrels) can be also extracted using denaturing agents. The peptide sequence that spans the membrane, or the transmembrane segment, is largely hydrophobic and can be visualized using the hydropathy plot. Depending on the number of transmembrane segments, transmembrane proteins can be classified as single-pass membrane proteins, or as multipass membrane proteins. Some other integral membrane proteins are called monotopic, meaning that they are also permanently attached to the membrane, b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytoplasm
The cytoplasm describes all the material within a eukaryotic or prokaryotic cell, enclosed by the cell membrane, including the organelles and excluding the nucleus in eukaryotic cells. The material inside the nucleus of a eukaryotic cell and contained within the nuclear membrane is termed the nucleoplasm. The main components of the cytoplasm are the cytosol (a gel-like substance), the cell's internal sub-structures, and various cytoplasmic inclusions. In eukaryotes the cytoplasm also includes the nucleus, and other membrane-bound organelles.The cytoplasm is about 80% water and is usually colorless. The submicroscopic ground cell substance, or cytoplasmic matrix, that remains after the exclusion of the cell organelles and particles is groundplasm. It is the hyaloplasm of light microscopy, a highly complex, polyphasic system in which all resolvable cytoplasmic elements are suspended, including the larger organelles such as the ribosomes, mitochondria, plant plasti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Disulfide Bond
In chemistry, a disulfide (or disulphide in British English) is a compound containing a functional group or the anion. The linkage is also called an SS-bond or sometimes a disulfide bridge and usually derived from two thiol groups. In inorganic chemistry, the anion appears in a few rare minerals, but the functional group has tremendous importance in biochemistry. Disulfide bridges formed between thiol groups in two cysteine residues are an important component of the tertiary and quaternary structure of proteins. Compounds of the form are usually called '' persulfides'' instead. Organic disulfides Structure Disulfides have a C–S–S–C dihedral angle approaching 90°. The S–S bond length is 2.03 Å in diphenyl disulfide, similar to that in elemental sulfur. Disulfides are usually symmetric but they can also be unsymmetric. Symmetrical disulfides are compounds of the formula . Most disulfides encountered in organosulfur chemistry are symmetrical disulfides. Unsy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Isoform
A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. (For that, see Proteoforms.) Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments (exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein. The discovery of isoforms could explain the discrepancy between the small number of protein coding regions of genes revealed by the human genome project and the large diversity of proteins seen in an organism: different proteins enc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |