|
ALAS2
Delta-aminolevulinate synthase 2 also known as ALAS2 is a protein that in humans is encoded by the ''ALAS2'' gene. ALAS2 is an aminolevulinic acid synthase. The product of this gene specifies an erythroid-specific mitochondrially located enzyme. The encoded protein catalyzes the first step in the heme biosynthetic pathway. Defects in this gene cause X-linked pyridoxine-responsive sideroblastic anemia. Alternatively spliced transcript variants encoding different isoforms have been identified. Its gene contains an IRE Ire or IRE may refer to: Ire * Extreme anger; intense fury * Irē, the Livonian name for Mazirbe, Latvia * A town in Oye, Nigeria * ''Ire'' (album), a 2015 album by the Australian metalcore band Parkway Drive * Ire (Iliad), a town mentioned in ... in its 5'-UTR region on which an IRP binds if the iron level is too low, thus inhibiting its translation. References Further reading * * * * * * * * * * * * * * * * * External links * Gene ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sideroblastic Anemia
Sideroblastic anemia, or sideroachrestic anemia, is a form of anemia in which the bone marrow produces ringed sideroblasts rather than healthy red blood cells (erythrocytes). In sideroblastic anemia, the body has iron available but cannot incorporate it into hemoglobin, which red blood cells need in order to transport oxygen efficiently. The disorder may be caused either by a genetic disorder or indirectly as part of myelodysplastic syndrome, which can develop into hematological malignancies (especially acute myeloid leukemia). Sideroblasts ('' sidero-'' + '' -blast'') are nucleated erythroblasts (precursors to mature red blood cells) with granules of iron accumulated in the mitochondria surrounding the nucleus. Normally, sideroblasts are present in the bone marrow, and enter the circulation after maturing into a normal erythrocyte. The presence of sideroblasts ''per se'' does not define sideroblastic anemia. Only the finding of ring (or ringed) sideroblasts characterizes sideroblas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aminolevulinic Acid Synthase
Aminolevulinic acid synthase (ALA synthase, ALAS, or delta-aminolevulinic acid synthase) is an enzyme () that catalyzes the synthesis of δ-aminolevulinic acid (ALA) the first common precursor in the biosynthesis of all tetrapyrroles such as hemes, cobalamins and chlorophylls. The reaction is as follows: :succinyl-CoA + glycine \rightleftharpoons δ-aminolevulinic acid + CoA + CO2 This enzyme is expressed in all non-plant eukaryotes and the α-class of proteobacteria and the reaction it catalyses is sometimes referred to as the Shemin pathway for ALA formation. Other organisms produce ALA through a three enzyme pathway known as the C5 pathway. ALA is synthesized through the condensation of glycine and succinyl-CoA. In humans, transcription of ALA synthase is tightly controlled by the presence of Fe2+-binding elements, to prevent accumulation of porphyrin intermediates in the absence of iron. There are two forms of ALA synthase in the body. One form is expressed in red blood cell ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heme
Heme, or haem (pronounced / hi:m/ ), is a precursor to hemoglobin, which is necessary to bind oxygen in the bloodstream. Heme is biosynthesized in both the bone marrow and the liver. In biochemical terms, heme is a coordination complex "consisting of an iron ion coordinated to a porphyrin acting as a tetradentate ligand, and to one or two axial ligands." The definition is loose, and many depictions omit the axial ligands. Among the metalloporphyrins deployed by metalloproteins as prosthetic groups, heme is one of the most widely used and defines a family of proteins known as hemoproteins. Hemes are most commonly recognized as components of hemoglobin, the red pigment in blood, but are also found in a number of other biologically important hemoproteins such as myoglobin, cytochromes, catalases, heme peroxidase, and endothelial nitric oxide synthase. The word ''haem'' is derived from Greek ''haima'' meaning "blood". Function Hemoproteins have diverse biological functions incl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Iron Response Element
In molecular biology, the iron response element or iron-responsive element (IRE) is a short conserved stem-loop which is bound by iron response proteins (IRPs, also named IRE-BP or IRBP). The IRE is found in UTRs (untranslated regions) of various mRNAs whose products are involved in iron metabolism. For example, the mRNA of ferritin (an iron storage protein) contains one IRE in its 5' UTR. When iron concentration is low, IRPs bind the IRE in the ferritin mRNA and cause reduced translation rates. In contrast, binding to multiple IREs in the 3' UTR of the transferrin receptor (involved in iron acquisition) leads to increased mRNA stability. Mechanism of action The two leading theories describe how iron probably interacts to impact posttranslational control of transcription. The classical theory suggests that IRPs, in the absence of iron, bind avidly to the mRNA IRE. When iron is present, it interacts with the protein to cause it to release the mRNA. For example, In high iro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitochondrion
A mitochondrion (; ) is an organelle found in the cells of most Eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used throughout the cell as a source of chemical energy. They were discovered by Albert von Kölliker in 1857 in the voluntary muscles of insects. The term ''mitochondrion'' was coined by Carl Benda in 1898. The mitochondrion is popularly nicknamed the "powerhouse of the cell", a phrase coined by Philip Siekevitz in a 1957 article of the same name. Some cells in some multicellular organisms lack mitochondria (for example, mature mammalian red blood cells). A large number of unicellular organisms, such as microsporidia, parabasalids and diplomonads, have reduced or transformed their mitochondria into other structures. One eukaryote, ''Monocercomonoides'', is known to have completely lost its mitochondria, and one multicellular organism, '' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
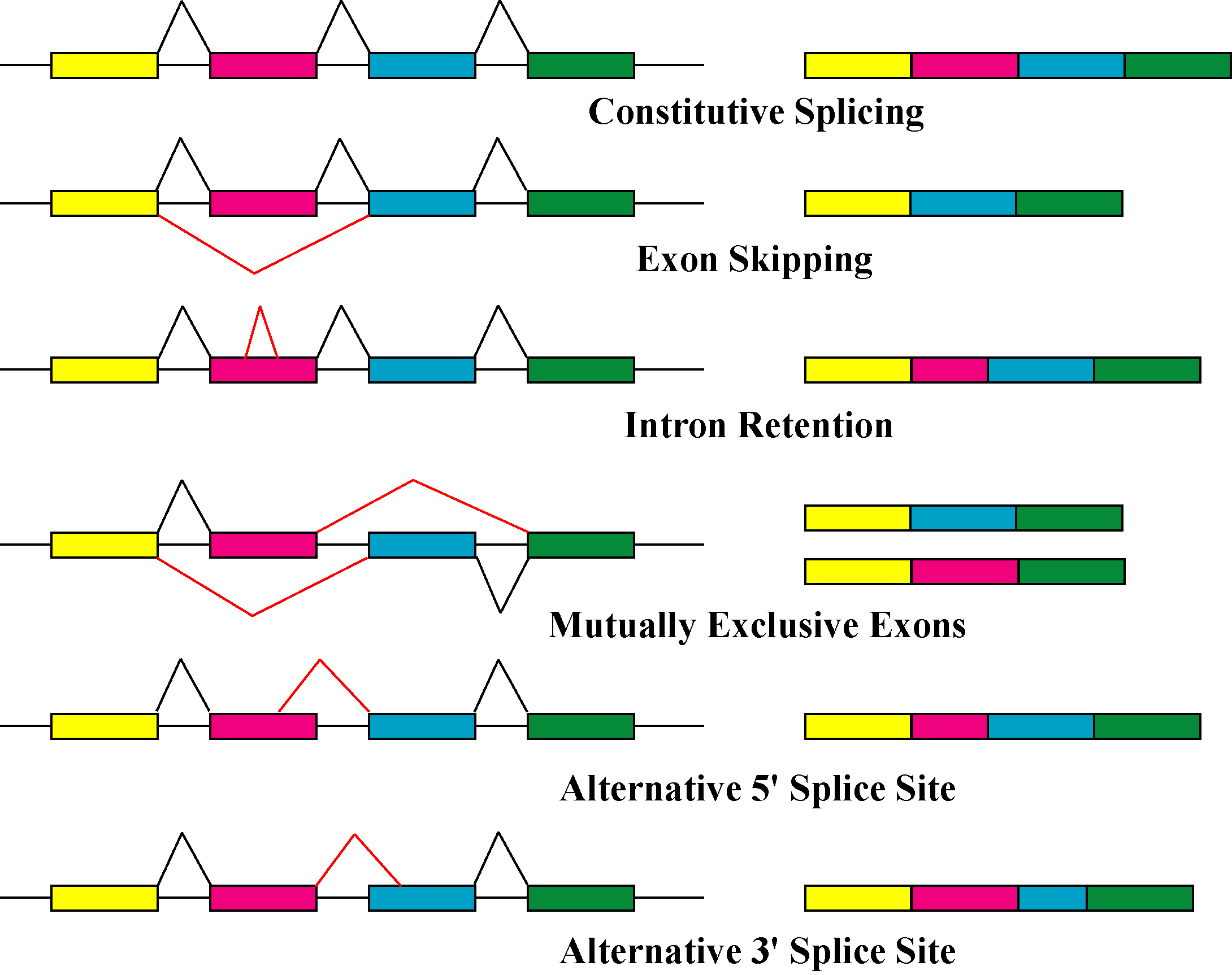
Alternative Splicing
Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. This means the exons are joined in different combinations, leading to different (alternative) mRNA strands. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions (see Figure). Biologically relevant alternative splicing occurs as a normal phenomenon in eukaryotes, where it increases the number of proteins that can be encoded by the genome. In humans, it is widely believed that ~95% of multi-exonic genes are alternatively spliced to produce functional alternative products from the same gene but many scientists believe that most o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Isoform
A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. (For that, see Proteoforms.) Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments ( exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein. The discovery of isoforms could explain the discrepancy between the small number of protein coding regions genes revealed by the human genome project and the large diversity of proteins seen in an organism: different ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |