|
ATG16L1
Autophagy related 16 like 1 is a protein that in humans is encoded by the ''ATG16L1'' gene. This protein is characterized as a subunit of the autophagy-related ATG12- ATG5/ATG16 complex and is essentially important for the LC3 ( ATG8) lipidation and autophagosome formation. This complex localizes to the membrane and is released just before or after autophagosome completion. Furthermore, ATG16L1 appears to have other autophagy-independent functions, e.g., intracellular membrane trafficking regulation and inflammation. Autophagy in general plays a crucial role in pathways leading to innate and adaptive immunity activation. That is why many autophagy-related proteins, including ATG16L1, their gene expression and its role in autoimmune diseases are studied in-depth nowadays. Function Autophagy is the major intracellular degradation system delivering cytoplasmic components to lysosomes, and it accounts for degradation of most long-lived proteins and some organelles. Cytoplasmic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Autophagy
Autophagy (or autophagocytosis; from the Greek language, Greek , , meaning "self-devouring" and , , meaning "hollow") is the natural, conserved degradation of the cell that removes unnecessary or dysfunctional components through a lysosome-dependent regulated mechanism. It allows the orderly degradation and recycling of cellular components. Although initially characterized as a primordial degradation pathway induced to protect against starvation, it has become increasingly clear that autophagy also plays a major role in the homeostasis of non-starved cells. Defects in autophagy have been linked to various human diseases, including neurodegeneration and cancer, and interest in modulating autophagy as a potential treatment for these diseases has grown rapidly. Four forms of autophagy have been identified: macroautophagy, microautophagy, chaperone-mediated autophagy (CMA), and crinophagy. In macroautophagy (the most thoroughly researched form of autophagy), cytoplasmic components ( ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Crohn's Disease
Crohn's disease is a type of inflammatory bowel disease (IBD) that may affect any segment of the gastrointestinal tract. Symptoms often include abdominal pain, diarrhea, fever, abdominal distension, and weight loss. Complications outside of the gastrointestinal tract may include anemia, skin rashes, arthritis, uveitis, inflammation of the eye, and fatigue (medical), fatigue. The skin rashes may be due to infections, as well as pyoderma gangrenosum or erythema nodosum. Bowel obstruction may occur as a complication of chronic inflammation, and those with the disease are at greater risk of colon cancer and small bowel cancer. Although the precise causes of Crohn's disease (CD) are unknown, it is believed to be caused by a combination of environmental, Immunity (medical), immune, and bacterial factors in genetically susceptible individuals. It results in a Immune-mediated inflammatory diseases, chronic inflammatory disorder, in which the body's immune system defends the gastrointesti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Autophagosome
An autophagosome is a spherical structure with double layer membranes. It is the key structure in macroautophagy, the intracellular degradation system for cytoplasmic contents (e.g., abnormal intracellular proteins, excess or damaged organelles, invading microorganisms). After formation, autophagosomes deliver cytoplasmic components to the lysosomes. The outer membrane of an autophagosome fuses with a lysosome to form an autolysosome. The lysosome's hydrolases degrade the autophagosome-delivered contents and its inner membrane. The formation of autophagosomes is regulated by genes that are well-conserved from yeast to higher eukaryotes. The nomenclature of these genes has differed from paper to paper, but it has been simplified in recent years. The gene families formerly known as APG, AUT, CVT, GSA, PAZ, and PDD are now unified as the ATG (AuTophaGy related) family. The size of autophagosomes varies between mammals and yeast. Yeast autophagosomes are about 500-900 nm, while m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
WD40 Repeat
The WD40 repeat (also known as the WD or beta-transducin repeat) is a short structural motif of approximately 40 amino acids, often terminating in a tryptophan-aspartic acid (W-D) dipeptide. Tandem copies of these repeats typically fold together to form a type of circular solenoid protein domain called the WD40 domain. Structure WD40 domain-containing proteins have 4 to 16 repeating units, all of which are thought to form a circularised beta-propeller structure (see figure to the right). The WD40 domain is composed of several repeats, a variable region of around 20 residues at the beginning followed by a more common repeated set of residues. These repeats typically form a four stranded anti-parallel beta sheet or blade. These blades come together to form a propeller with the most common being a 7 bladed beta propeller. The blades interlock so that the last beta strand of one repeat forms with the first three of the next repeat to form the 3D blade structure. Function WD40-re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ATG5
Autophagy protein 5 (ATG5) is a protein that, in humans, is encoded by the ''ATG5'' gene located on chromosome 6. It is an E3 ubi autophagic cell death. ATG5 is a key protein involved in the extension of the phagophoric membrane in autophagic vesicles. It is activated by ATG7 and forms a complex with ATG12 and ATG16L1. This complex is necessary for LC3-I (microtubule-associated proteins 1A/1B light chain 3B) conjugation to PE (phosphatidylethanolamine) to form LC3-II (LC3-phosphatidylethanolamine conjugate). ATG5 can also act as a pro-apoptotic molecule targeted to the mitochondria. Under low levels of DNA damage, ATG5 can translocate to the nucleus and interact with survivin. ATG5 is known to be regulated via various stress induced transcription factors and protein kinases. Structure ATG5 comprises three domains: a ubiquitin-like N-terminal domain (UblA), a helix-rich domain (HR) and a ubiquitin-like C-terminal domain (UblB). The three domains are connected by two linke ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metabolic reactions, DNA replication, Cell signaling, responding to stimuli, providing Cytoskeleton, structure to cells and Fibrous protein, organisms, and Intracellular transport, transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the Nucleic acid sequence, nucleotide sequence of their genes, and which usually results in protein folding into a specific Protein structure, 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called pep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
An alpha helix (or α-helix) is a sequence of amino acids in a protein that are twisted into a coil (a helix). The alpha helix is the most common structural arrangement in the Protein secondary structure, secondary structure of proteins. It is also the most extreme type of local structure, and it is the local structure that is most easily predicted from a sequence of amino acids. The alpha helix has a right-handed helix conformation in which every backbone amino, N−H group hydrogen bonds to the backbone carbonyl, C=O group of the amino acid that is four residue (biochemistry), residues earlier in the protein sequence. Other names The alpha helix is also commonly called a: * Pauling–Corey–Branson α-helix (from the names of three scientists who described its structure) * 3.613-helix because there are 3.6 amino acids in one ring, with 13 atoms being involved in the ring formed by the hydrogen bond (starting with amidic hydrogen and ending with carbonyl oxygen) Discovery ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A. The addition of ubiquitin to a substrate protein is called ubiquitylation (or ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the 26S proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide bond, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
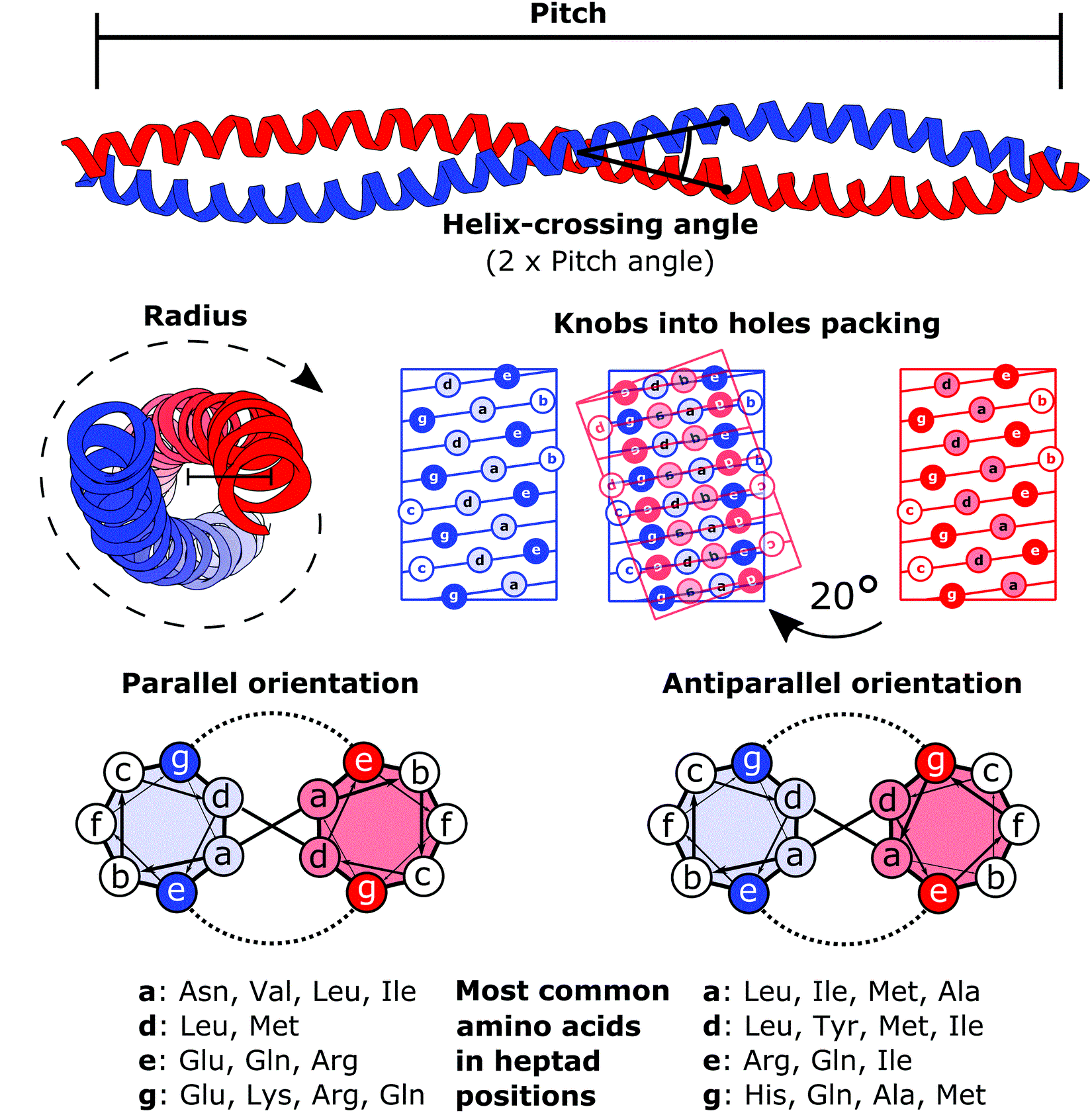
Coiled Coil
A coiled coil is a structural motif in proteins in which two to seven alpha-helices are coiled together like the strands of a rope. ( Dimers and trimers are the most common types.) They have been found in roughly 5-10% of proteins and have a variety of functions. They are one of the most widespread motifs found in protein-protein interactions. To aid protein study, several tools have been developed to predict coiled-coils in protein structures. Many coiled coil-type proteins are involved in important biological functions, such as the regulation of gene expression — e.g., transcription factors. Notable examples are the oncoproteins c-Fos and c-Jun, as well as the muscle protein tropomyosin. Discovery The possibility of coiled coils for α-keratin was initially somewhat controversial. Linus Pauling and Francis Crick independently came to the conclusion that this was possible at about the same time. In the summer of 1952, Pauling visited the laboratory in England where C ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology (physical form and structure), its developmental processes, its biochemical and physiological properties, and its behavior. An organism's phenotype results from two basic factors: the expression of an organism's genetic code (its genotype) and the influence of environmental factors. Both factors may interact, further affecting the phenotype. When two or more clearly different phenotypes exist in the same population of a species, the species is called polymorphic. A well-documented example of polymorphism is Labrador Retriever coloring; while the coat color depends on many genes, it is clearly seen in the environment as yellow, black, and brown. Richard Dawkins in 1978 and again in his 1982 book '' The Extended Phenotype'' suggested that one can regard bird nests and other built structures such as caddisfly larva cases and beaver dams ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymorphism (biology)
In biology, polymorphism is the occurrence of two or more clearly different morphs or forms, also referred to as alternative '' phenotypes'', in the population of a species. To be classified as such, morphs must occupy the same habitat at the same time and belong to a panmictic population (one with random mating). Ford E.B. 1965. ''Genetic polymorphism''. Faber & Faber, London. Put simply, polymorphism is when there are two or more possibilities of a trait on a gene. For example, there is more than one possible trait in terms of a jaguar's skin colouring; they can be light morph or dark morph. Due to having more than one possible variation for this gene, it is termed 'polymorphism'. However, if the jaguar has only one possible trait for that gene, it would be termed "monomorphic". For example, if there was only one possible skin colour that a jaguar could have, it would be termed monomorphic. The term polyphenism can be used to clarify that the different forms arise from the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |