Linkage analysis on:
[Wikipedia]
[Google]
[Amazon]
Genetic linkage is the tendency of
 A linkage map (also known as a genetic map) is a table for a species or experimental population that shows the position of its known genes or
A linkage map (also known as a genetic map) is a table for a species or experimental population that shows the position of its known genes or
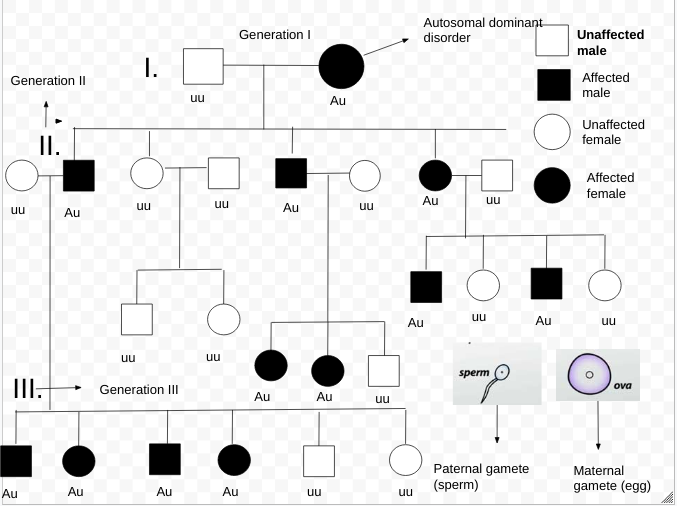
Briefly, it works as follows: # Establish a Pedigree chart, pedigree # Make a number of estimates of recombination frequency # Calculate a LOD score for each estimate # The estimate with the highest LOD score will be considered the best estimate The LOD score is calculated as follows: : NR denotes the number of non-recombinant offspring, and R denotes the number of recombinant offspring. The reason 0.5 is used in the denominator is that any alleles that are completely unlinked (e.g. alleles on separate chromosomes) have a 50% chance of recombination, due to independent assortment. ''θ'' is the recombinant fraction, i.e. the fraction of births in which recombination has happened between the studied genetic marker and the putative gene associated with the disease. Thus, it is equal to . By convention, a LOD score greater than 3.0 is considered evidence for linkage, as it indicates 1000 to 1 odds that the linkage being observed did not occur by chance. On the other hand, a LOD score less than −2.0 is considered evidence to exclude linkage. Although it is very unlikely that a LOD score of 3 would be obtained from a single pedigree, the mathematical properties of the test allow data from a number of pedigrees to be combined by summing their LOD scores. A LOD score of 3 translates to a ''p''-value of approximately 0.05, and no multiple testing correction (e.g.
 Their experiment revealed linkage between the ''P'' and ''L'' alleles and the ''p'' and ''l'' alleles. The frequency of ''P'' occurring together with ''L'' and with ''p'' occurring together with ''l'' is greater than that of the recombinant ''Pl'' and ''pL''. The recombination frequency is more difficult to compute in an F2 cross than a backcross, but the lack of fit between observed and expected numbers of progeny in the above table indicate it is less than 50%.
The progeny in this case received two dominant alleles linked on one chromosome (referred to as coupling or cis arrangement). However, after crossover, some progeny could have received one parental chromosome with a dominant allele for one trait (e.g. Purple) linked to a recessive allele for a second trait (e.g. round) with the opposite being true for the other parental chromosome (e.g. red and Long). This is referred to as repulsion or a trans arrangement. The
Their experiment revealed linkage between the ''P'' and ''L'' alleles and the ''p'' and ''l'' alleles. The frequency of ''P'' occurring together with ''L'' and with ''p'' occurring together with ''l'' is greater than that of the recombinant ''Pl'' and ''pL''. The recombination frequency is more difficult to compute in an F2 cross than a backcross, but the lack of fit between observed and expected numbers of progeny in the above table indicate it is less than 50%.
The progeny in this case received two dominant alleles linked on one chromosome (referred to as coupling or cis arrangement). However, after crossover, some progeny could have received one parental chromosome with a dominant allele for one trait (e.g. Purple) linked to a recessive allele for a second trait (e.g. round) with the opposite being true for the other parental chromosome (e.g. red and Long). This is referred to as repulsion or a trans arrangement. The
DNA sequences
A nucleic acid sequence is a succession of bases signified by a series of a set of five different letters that indicate the order of nucleotides forming alleles within a DNA (using GACT) or RNA (GACU) molecule. By convention, sequences are us ...
that are close together on a chromosome
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins are ...
to be inherited together during the meiosis
Meiosis (; , since it is a reductional division) is a special type of cell division of germ cells in sexually-reproducing organisms that produces the gametes, such as sperm or egg cells. It involves two rounds of division that ultimately r ...
phase of sexual reproduction
Sexual reproduction is a type of reproduction that involves a complex life cycle in which a gamete ( haploid reproductive cells, such as a sperm or egg cell) with a single set of chromosomes combines with another gamete to produce a zygote th ...
. Two genetic markers A genetic marker is a gene or DNA sequence with a known location on a chromosome that can be used to identify individuals or species. It can be described as a variation (which may arise due to mutation or alteration in the genomic loci) that can ...
that are physically near to each other are unlikely to be separated onto different chromatids
A chromatid (Greek ''khrōmat-'' 'color' + ''-id'') is one half of a duplicated chromosome. Before replication, one chromosome is composed of one DNA molecule. In replication, the DNA molecule is copied, and the two molecules are known as chro ...
during chromosomal crossover, and are therefore said to be more ''linked'' than markers that are far apart. In other words, the nearer two genes are on a chromosome, the lower the chance of recombination between them, and the more likely they are to be inherited together. Markers on different chromosomes are perfectly ''unlinked'', although the penetrance
Penetrance in genetics is the proportion of individuals carrying a particular variant (or allele) of a gene (the genotype) that also express an associated trait (the phenotype). In medical genetics, the penetrance of a disease-causing mutation is t ...
of potentially deleterious alleles may be influenced by the presence of other alleles, and these other alleles may be located on other chromosomes than that on which a particular potentially deleterious allele is located.
Genetic linkage is the most prominent exception to Gregor Mendel
Gregor Johann Mendel, OSA (; cs, Řehoř Jan Mendel; 20 July 1822 – 6 January 1884) was a biologist, meteorologist, mathematician, Augustinian friar and abbot of St. Thomas' Abbey in Brünn (''Brno''), Margraviate of Moravia. Mendel was ...
's Law of Independent Assortment
Mendelian inheritance (also known as Mendelism) is a type of biological inheritance following the principles originally proposed by Gregor Mendel in 1865 and 1866, re-discovered in 1900 by Hugo de Vries and Carl Correns, and later populariz ...
. The first experiment to demonstrate linkage was carried out in 1905. At the time, the reason why certain traits tend to be inherited together was unknown. Later work revealed that genes are physical structures related by physical distance.
The typical unit of genetic linkage is the centimorgan
In genetics, a centimorgan (abbreviated cM) or map unit (m.u.) is a unit for measuring genetic linkage. It is defined as the distance between chromosome positions (also termed loci or markers) for which the expected average number of intervening ...
(cM). A distance of 1 cM between two markers means that the markers are separated to different chromosomes on average once per 100 meiotic product, thus once per 50 meioses.
Discovery
Gregor Mendel
Gregor Johann Mendel, OSA (; cs, Řehoř Jan Mendel; 20 July 1822 – 6 January 1884) was a biologist, meteorologist, mathematician, Augustinian friar and abbot of St. Thomas' Abbey in Brünn (''Brno''), Margraviate of Moravia. Mendel was ...
's Law of Independent Assortment
Mendelian inheritance (also known as Mendelism) is a type of biological inheritance following the principles originally proposed by Gregor Mendel in 1865 and 1866, re-discovered in 1900 by Hugo de Vries and Carl Correns, and later populariz ...
states that every trait is inherited independently of every other trait. But shortly after Mendel's work was rediscovered, exceptions to this rule were found. In 1905, the British
British may refer to:
Peoples, culture, and language
* British people, nationals or natives of the United Kingdom, British Overseas Territories, and Crown Dependencies.
** Britishness, the British identity and common culture
* British English, ...
geneticists William Bateson
William Bateson (8 August 1861 – 8 February 1926) was an English biologist who was the first person to use the term genetics to describe the study of heredity, and the chief populariser of the ideas of Gregor Mendel following their rediscove ...
, Edith Rebecca Saunders and Reginald Punnett
Reginald Crundall Punnett FRS (; 20 June 1875 – 3 January 1967) was a British geneticist who co-founded, with William Bateson, the ''Journal of Genetics'' in 1910. Punnett is probably best remembered today as the creator of the Punnett ...
cross-bred pea plants in experiments similar to Mendel's. They were interested in trait inheritance in the sweet pea and were studying two genes—the gene for flower colour (''P'', purple, and ''p'', red) and the gene affecting the shape of pollen grains (''L'', long, and ''l'', round). They crossed the pure lines ''PPLL'' and ''ppll'' and then self-crossed the resulting ''PpLl'' lines.
According to Mendelian genetics
Mendelian inheritance (also known as Mendelism) is a type of biological inheritance following the principles originally proposed by Gregor Mendel in 1865 and 1866, re-discovered in 1900 by Hugo de Vries and Carl Correns, and later populari ...
, the expected phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology or physical form and structure, its developmental processes, its biochemical and physiological pr ...
s would occur in a 9:3:3:1 ratio of PL:Pl:pL:pl. To their surprise, they observed an increased frequency of PL and pl and a decreased frequency of Pl and pL:
Their experiment revealed linkage between the ''P'' and ''L'' alleles and the ''p'' and ''l'' alleles. The frequency of ''P'' occurring together with ''L'' and ''p'' occurring together with ''l'' is greater than that of the recombinant ''Pl'' and ''pL''. The recombination frequency is more difficult to compute in an F2 cross than a backcross, but the lack of fit between observed and expected numbers of progeny in the above table indicate it is less than 50%. This indicated that two factors interacted in some way to create this difference by masking the appearance of the other two phenotypes. This led to the conclusion that some traits are related to each other because of their near proximity to each other on a chromosome.
The understanding of linkage was expanded by the work of Thomas Hunt Morgan
Thomas Hunt Morgan (September 25, 1866 – December 4, 1945) was an American evolutionary biologist, geneticist, embryologist, and science author who won the Nobel Prize in Physiology or Medicine in 1933 for discoveries elucidating the role that ...
. Morgan's observation that the amount of crossing over between linked genes differs led to the idea that crossover frequency might indicate the distance separating genes on the chromosome
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins are ...
. The centimorgan
In genetics, a centimorgan (abbreviated cM) or map unit (m.u.) is a unit for measuring genetic linkage. It is defined as the distance between chromosome positions (also termed loci or markers) for which the expected average number of intervening ...
, which expresses the frequency of crossing over, is named in his honour.
Linkage map
genetic markers A genetic marker is a gene or DNA sequence with a known location on a chromosome that can be used to identify individuals or species. It can be described as a variation (which may arise due to mutation or alteration in the genomic loci) that can ...
relative to each other in terms of recombination frequency, rather than a specific physical distance along each chromosome. Linkage maps were first developed by Alfred Sturtevant
Alfred Henry Sturtevant (November 21, 1891 – April 5, 1970) was an American geneticist. Sturtevant constructed the first genetic map of a chromosome in 1911. Throughout his career he worked on the organism ''Drosophila melanogaster'' with ...
, a student of Thomas Hunt Morgan
Thomas Hunt Morgan (September 25, 1866 – December 4, 1945) was an American evolutionary biologist, geneticist, embryologist, and science author who won the Nobel Prize in Physiology or Medicine in 1933 for discoveries elucidating the role that ...
.
A linkage map is a map based on the frequencies of recombination between markers during crossover
Crossover may refer to:
Entertainment
Albums and songs
* ''Cross Over'' (Dan Peek album)
* ''Crossover'' (Dirty Rotten Imbeciles album), 1987
* ''Crossover'' (Intrigue album)
* ''Crossover'' (Hitomi Shimatani album)
* ''Crossover'' (Yoshino ...
of homologous chromosome
A couple of homologous chromosomes, or homologs, are a set of one maternal and one paternal chromosome that pair up with each other inside a cell during fertilization. Homologs have the same genes in the same loci where they provide points alon ...
s. The greater the frequency of recombination (segregation) between two genetic markers, the further apart they are assumed to be. Conversely, the lower the frequency of recombination between the markers, the smaller the physical distance between them. Historically, the markers originally used were detectable phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology or physical form and structure, its developmental processes, its biochemical and physiological pr ...
s (enzyme production, eye colour) derived from coding DNA
The coding region of a gene, also known as the coding sequence (CDS), is the portion of a gene's DNA or RNA that codes for protein. Studying the length, composition, regulation, splicing, structures, and functions of coding regions compared to non ...
sequences; eventually, confirmed or assumed noncoding DNA
Non-coding DNA (ncDNA) sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA is transcribed into functional non-coding RNA molecules (e.g. transfer RNA, microRNA, piRNA, ribosomal RNA, and r ...
sequences such as microsatellites
A microsatellite is a tract of repetitive DNA in which certain DNA motifs (ranging in length from one to six or more base pairs) are repeated, typically 5–50 times. Microsatellites occur at thousands of locations within an organism's genome. ...
or those generating restriction fragment length polymorphisms ( RFLPs) have been used.
Linkage maps help researchers to locate other markers, such as other genes by testing for genetic linkage of the already known markers. In the early stages of developing a linkage map, the data are used to assemble linkage groups, a set of genes which are known to be linked. As knowledge advances, more markers can be added to a group, until the group covers an entire chromosome. For well-studied organisms the linkage groups correspond one-to-one with the chromosomes.
A linkage map is not a physical map (such as a radiation reduced hybrid map) or gene map
Gene maps help describe the spatial arrangement of genes on a chromosome. Genes are designated to a specific location on a chromosome known as the locus and can be used as molecular markers to find the distance between other genes on a chromosom ...
.
Linkage analysis
Linkage analysis is a genetic method that searches for chromosomal segments that cosegregate with the ailment phenotype through families. It can be used to map genes for both binary and quantitative traits. Linkage analysis may be either parametric (if we know the relationship between phenotypic and genetic similarity) or non-parametric. Parametric linkage analysis is the traditional approach, whereby the probability that a gene important for a disease is linked to a genetic marker is studied through the LOD score, which assesses the probability that a given pedigree, where the disease and the marker are cosegregating, is due to the existence of linkage (with a given linkage value) or to chance. Non-parametric linkage analysis, in turn, studies the probability of an allele beingidentical by descent
A DNA segment is identical by state (IBS) in two or more individuals if they have identical nucleotide sequences in this segment. An IBS segment is identical by descent (IBD) in two or more individuals if they have inherited it from a common an ...
with itself.

Parametric linkage analysis
The LOD score (logarithm (base 10) of odds), developed byNewton Morton
Newton Ennis Morton (21 December 1929 – 7 February 2018) was an American population geneticist and one of the founders of the field of genetic epidemiology.
Early life and education
Morton was born in Camden, New Jersey. When he was three ...
, is a statistical test often used for linkage analysis in human, animal, and plant populations. The LOD score compares the likelihood of obtaining the test data if the two loci are indeed linked, to the likelihood of observing the same data purely by chance. Positive LOD scores favour the presence of linkage, whereas negative LOD scores indicate that linkage is less likely. Computerised LOD score analysis is a simple way to analyse complex family pedigrees in order to determine the linkage between Mendelian
Mendelian inheritance (also known as Mendelism) is a type of biological inheritance following the principles originally proposed by Gregor Mendel in 1865 and 1866, re-discovered in 1900 by Hugo de Vries and Carl Correns, and later popularize ...
traits (or between a trait and a marker, or two markers).
The method is described in greater detail by Strachan and ReaBriefly, it works as follows: # Establish a Pedigree chart, pedigree # Make a number of estimates of recombination frequency # Calculate a LOD score for each estimate # The estimate with the highest LOD score will be considered the best estimate The LOD score is calculated as follows: : NR denotes the number of non-recombinant offspring, and R denotes the number of recombinant offspring. The reason 0.5 is used in the denominator is that any alleles that are completely unlinked (e.g. alleles on separate chromosomes) have a 50% chance of recombination, due to independent assortment. ''θ'' is the recombinant fraction, i.e. the fraction of births in which recombination has happened between the studied genetic marker and the putative gene associated with the disease. Thus, it is equal to . By convention, a LOD score greater than 3.0 is considered evidence for linkage, as it indicates 1000 to 1 odds that the linkage being observed did not occur by chance. On the other hand, a LOD score less than −2.0 is considered evidence to exclude linkage. Although it is very unlikely that a LOD score of 3 would be obtained from a single pedigree, the mathematical properties of the test allow data from a number of pedigrees to be combined by summing their LOD scores. A LOD score of 3 translates to a ''p''-value of approximately 0.05, and no multiple testing correction (e.g.
Bonferroni correction
In statistics, the Bonferroni correction is a method to counteract the multiple comparisons problem.
Background
The method is named for its use of the Bonferroni inequalities.
An extension of the method to confidence intervals was proposed by Ol ...
) is required.
Limitations
Linkage analysis has a number of methodological and theoretical limitations that can significantly increase the type-1 error rate and reduce the power to map human quantitative trait loci (QTL). While linkage analysis was successfully used to identify genetic variants that contribute to rare disorders such asHuntington disease
Huntington's disease (HD), also known as Huntington's chorea, is a neurodegenerative disease that is mostly inherited. The earliest symptoms are often subtle problems with mood or mental abilities. A general lack of coordination and an unst ...
, it did not perform that well when applied to more common disorders such as heart disease or different forms of cancer
Cancer is a group of diseases involving abnormal cell growth with the potential to invade or spread to other parts of the body. These contrast with benign tumors, which do not spread. Possible signs and symptoms include a lump, abnormal b ...
. An explanation for this is that the genetic mechanisms affecting common disorders are different from those causing some rare disorders.
Recombination frequency
Recombination frequency is a measure of genetic linkage and is used in the creation of a genetic linkage map. Recombination frequency (''θ'') is the frequency with which a single chromosomal crossover will take place between twogene
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b ...
s during meiosis
Meiosis (; , since it is a reductional division) is a special type of cell division of germ cells in sexually-reproducing organisms that produces the gametes, such as sperm or egg cells. It involves two rounds of division that ultimately r ...
. A centimorgan
In genetics, a centimorgan (abbreviated cM) or map unit (m.u.) is a unit for measuring genetic linkage. It is defined as the distance between chromosome positions (also termed loci or markers) for which the expected average number of intervening ...
(cM) is a unit that describes a recombination frequency of 1%. In this way we can measure the genetic distance between two loci, based upon their recombination frequency. This is a good estimate of the real distance. Double crossovers would turn into no recombination. In this case we cannot tell if crossovers took place. If the loci we're analysing are very close (less than 7 cM) a double crossover is very unlikely. When distances become higher, the likelihood of a double crossover increases. As the likelihood of a double crossover increases one could systematically underestimate the genetic distance between two loci, unless one used an appropriate mathematical model.
During meiosis, chromosomes assort randomly into gamete
A gamete (; , ultimately ) is a haploid cell that fuses with another haploid cell during fertilization in organisms that reproduce sexually. Gametes are an organism's reproductive cells, also referred to as sex cells. In species that produce ...
s, such that the segregation of allele
An allele (, ; ; modern formation from Greek ἄλλος ''állos'', "other") is a variation of the same sequence of nucleotides at the same place on a long DNA molecule, as described in leading textbooks on genetics and evolution.
::"The chro ...
s of one gene is independent of alleles of another gene. This is stated in Mendel's Second Law and is known as the law of independent assortment. The law of independent assortment always holds true for genes that are located on different chromosomes, but for genes that are on the same chromosome, it does not always hold true.
As an example of independent assortment, consider the crossing of the pure-bred homozygote
Zygosity (the noun, zygote, is from the Greek "yoked," from "yoke") () is the degree to which both copies of a chromosome or gene have the same genetic sequence. In other words, it is the degree of similarity of the alleles in an organism.
Mo ...
parental strain with genotype ''AABB'' with a different pure-bred strain with genotype ''aabb''. A and a and B and b represent the alleles of genes A and B. Crossing these homozygous parental strains will result in F1 generation offspring that are double heterozygotes
Zygosity (the noun, zygote, is from the Greek "yoked," from "yoke") () is the degree to which both copies of a chromosome or gene have the same genetic sequence. In other words, it is the degree of similarity of the alleles in an organism.
Mo ...
with genotype AaBb. The F1 offspring AaBb produces gametes that are ''AB'', ''Ab'', ''aB'', and ''ab'' with equal frequencies (25%) because the alleles of gene A assort independently of the alleles for gene B during meiosis. Note that 2 of the 4 gametes (50%)—''Ab'' and ''aB''—were not present in the parental generation. These gametes represent recombinant gametes. Recombinant gametes are those gametes that differ from both of the haploid gametes that made up the original diploid cell. In this example, the recombination frequency is 50% since 2 of the 4 gametes were recombinant gametes.
The recombination frequency will be 50% when two genes are located on different chromosomes
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins are ...
or when they are widely separated on the same chromosome. This is a consequence of independent assortment.
When two genes are close together on the same chromosome, they do not assort independently and are said to be linked. Whereas genes located on different chromosomes assort independently and have a recombination frequency of 50%, linked genes have a recombination frequency that is less than 50%.
As an example of linkage, consider the classic experiment by William Bateson
William Bateson (8 August 1861 – 8 February 1926) was an English biologist who was the first person to use the term genetics to describe the study of heredity, and the chief populariser of the ideas of Gregor Mendel following their rediscove ...
and Reginald Punnett
Reginald Crundall Punnett FRS (; 20 June 1875 – 3 January 1967) was a British geneticist who co-founded, with William Bateson, the ''Journal of Genetics'' in 1910. Punnett is probably best remembered today as the creator of the Punnett ...
. They were interested in trait inheritance in the sweet pea and were studying two genes—the gene for flower colour (''P'', purple, and ''p'', red) and the gene affecting the shape of pollen grains (''L'', long, and ''l'', round). They crossed the pure lines ''PPLL'' and ''ppll'' and then self-crossed the resulting ''PpLl'' lines. According to Mendelian genetics
Mendelian inheritance (also known as Mendelism) is a type of biological inheritance following the principles originally proposed by Gregor Mendel in 1865 and 1866, re-discovered in 1900 by Hugo de Vries and Carl Correns, and later populari ...
, the expected phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology or physical form and structure, its developmental processes, its biochemical and physiological pr ...
s would occur in a 9:3:3:1 ratio of PL:Pl:pL:pl. To their surprise, they observed an increased frequency of PL and pl and a decreased frequency of Pl and pL (see table below).
phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology or physical form and structure, its developmental processes, its biochemical and physiological pr ...
here would still be purple and long but a test cross of this individual with the recessive parent would produce progeny with much greater proportion of the two crossover phenotypes. While such a problem may not seem likely from this example, unfavourable repulsion linkages do appear when breeding for disease resistance in some crops.
The two possible arrangements, cis and trans, of alleles in a double heterozygote are referred to as gametic phase In genetics, a gametic phase represents the original allelic combinations that a diploid individual inherits from both parents. It is therefore a particular association of alleles at different loci on the same chromosome. Gametic phase is influence ...
s, and ''phasing'' is the process of determining which of the two is present in a given individual.
When two genes are located on the same chromosome, the chance of a crossover
Crossover may refer to:
Entertainment
Albums and songs
* ''Cross Over'' (Dan Peek album)
* ''Crossover'' (Dirty Rotten Imbeciles album), 1987
* ''Crossover'' (Intrigue album)
* ''Crossover'' (Hitomi Shimatani album)
* ''Crossover'' (Yoshino ...
producing recombination between the genes is related to the distance between the two genes. Thus, the use of recombination frequencies has been used to develop linkage maps or genetic maps.
However, it is important to note that recombination frequency tends to underestimate the distance between two linked genes. This is because as the two genes are located farther apart, the chance of double or even number of crossovers between them also increases. Double or even number of crossovers between the two genes results in them being cosegregated to the same gamete, yielding a parental progeny instead of the expected recombinant progeny. As mentioned above, the Kosambi and Haldane transformations attempt to correct for multiple crossovers.
Linkage of genetic sites within a gene
In the early 1950s the prevailing view was that the genes in achromosome
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins are ...
are discrete entities, indivisible by genetic recombination and arranged like beads on a string. During 1955 to 1959, Benzer performed genetic recombination experiments using rII mutants of bacteriophage T4
Escherichia virus T4 is a species of bacteriophages that infect ''Escherichia coli'' bacteria. It is a double-stranded DNA virus in the subfamily '' Tevenvirinae'' from the family Myoviridae. T4 is capable of undergoing only a lytic lifecycle ...
. He found that, on the basis of recombination tests, the sites of mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA replication, DNA or viral repl ...
could be mapped in a linear order. This result provided evidence for the key idea that the gene has a linear structure equivalent to a length of DNA with many sites that can independently mutate.
Edgar et al. performed mapping experiments with r mutants of bacteriophage T4 showing that recombination frequencies between rII mutants are not strictly additive. The recombination frequency from a cross of two rII mutants (a x d) is usually less than the sum of recombination frequencies for adjacent internal sub-intervals (a x b) + (b x c) + (c x d). Although not strictly additive, a systematic relationship was observed that likely reflects the underlying molecular mechanism of genetic recombination.
Variation of recombination frequency
While recombination of chromosomes is an essential process during meiosis, there is a large range of frequency of cross overs across organisms and within species. Sexually dimorphic rates of recombination are termed heterochiasmy, and are observed more often than a common rate between male and females. In mammals, females often have a higher rate of recombination compared to males. It is theorised that there are unique selections acting or meiotic drivers which influence the difference in rates. The difference in rates may also reflect the vastly different environments and conditions of meiosis in oogenesis and spermatogenesis.Genes affecting recombination frequency
Mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA replication, DNA or viral repl ...
s in gene
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b ...
s that encode proteins involved in the processing of DNA often affect recombination frequency. In bacteriophage T4
Escherichia virus T4 is a species of bacteriophages that infect ''Escherichia coli'' bacteria. It is a double-stranded DNA virus in the subfamily '' Tevenvirinae'' from the family Myoviridae. T4 is capable of undergoing only a lytic lifecycle ...
, mutations that reduce expression of the replicative DNA polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create ...
ene product 43 (gp43)increase recombination (decrease linkage) several fold.Bernstein H. The effect on recombination of mutational defects in the DNA-polymerase and deoxycytidylate hydroxymethylase of phage T4D. Genetics. 1967;56(4):755-769Berger H, Warren AJ, Fry KE. Variations in genetic recombination due to amber mutations in T4D bacteriophage. J Virol. 1969;3(2):171-175. doi:10.1128/JVI.3.2.171-175.1969 The increase in recombination may be due to replication errors by the defective DNA polymerase that are themselves recombination events such as template switches, i.e. copy choice recombination events. Recombination is also increased by mutations that reduce the expression of DNA ligase
DNA ligase is a specific type of enzyme, a ligase, () that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond. It plays a role in repairing single-strand breaks in duplex DNA in living orga ...
(gp30)Bernstein H. Repair and recombination in phage T4. I. Genes affecting recombination. Cold Spring Harb Symp Quant Biol. 1968;33:325-331. doi:10.1101/sqb.1968.033.01.037 and dCMP hydroxymethylase (gp42), two enzymes employed in DNA synthesis
DNA synthesis is the natural or artificial creation of deoxyribonucleic acid (DNA) molecules. DNA is a macromolecule made up of nucleotide units, which are linked by covalent bonds and hydrogen bonds, in a repeating structure. DNA synthesis occurs ...
.
Recombination is reduced (linkage increased) by mutations in genes that encode proteins with nuclease functions (gp46 and gp47) and a DNA-binding protein
DNA-binding proteins are proteins that have DNA-binding domains and thus have a specific or general affinity for single- or double-stranded DNA. Sequence-specific DNA-binding proteins generally interact with the major groove of B-DNA, becaus ...
(gp32) Mutation in the bacteriophage uvsX gene also substantially reduces recombination. The uvsX gene is analogous to the well studied ''recA
RecA is a 38 kilodalton protein essential for the repair and maintenance of DNA. A RecA structural and functional homolog has been found in every species in which one has been seriously sought and serves as an archetype for this class of homolog ...
'' gene of ''Escherichia coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus '' Esc ...
'' that plays a central role in recombination.Fujisawa H, Yonesaki T, Minagawa T. Sequence of the T4 recombination gene, uvsX, and its comparison with that of the recA gene of Escherichia coli. Nucleic Acids Res. 1985;13(20):7473-7481. doi:10.1093/nar/13.20.7473
Meiosis indicators
With very large pedigrees or with very dense genetic marker data, such as from whole-genome sequencing, it is possible to precisely locate recombinations. With this type of genetic analysis, a meiosis indicator is assigned to each position of the genome for eachmeiosis
Meiosis (; , since it is a reductional division) is a special type of cell division of germ cells in sexually-reproducing organisms that produces the gametes, such as sperm or egg cells. It involves two rounds of division that ultimately r ...
in a pedigree. The indicator indicates which copy of the parental chromosome contributes to the transmitted gamete at that position. For example, if the allele from the 'first' copy of the parental chromosome is transmitted, a '0' might be assigned to that meiosis. If the allele from the 'second' copy of the parental chromosome is transmitted, a '1' would be assigned to that meiosis. The two alleles in the parent came, one each, from two grandparents. These indicators are then used to determine identical-by-descent (IBD) states or inheritance states, which are in turn used to identify genes responsible for diseases.
See also
*Centimorgan
In genetics, a centimorgan (abbreviated cM) or map unit (m.u.) is a unit for measuring genetic linkage. It is defined as the distance between chromosome positions (also termed loci or markers) for which the expected average number of intervening ...
* Genetic association
Genetic association is when one or more genotypes within a population co-occur with a phenotypic trait more often than would be expected by chance occurrence.
Studies of genetic association aim to test whether single-locus alleles or genotype fre ...
* Genetic epidemiology
* Genome-wide association study
In genomics, a genome-wide association study (GWA study, or GWAS), also known as whole genome association study (WGA study, or WGAS), is an observational study of a genome-wide set of genetic variants in different individuals to see if any vari ...
* Identity by descent
A DNA segment is identical by state (IBS) in two or more individuals if they have identical nucleotide sequences in this segment. An IBS segment is identical by descent (IBD) in two or more individuals if they have inherited it from a common a ...
* Lander–Green algorithm
The Lander–Green algorithm is an algorithm, due to Eric Lander and Philip Green for computing the likelihood of observed genotype data given a pedigree. It is appropriate for relatively small pedigrees and a large number of markers. It is used in ...
* Linkage disequilibrium
In population genetics, linkage disequilibrium (LD) is the non-random association of alleles at different loci in a given population. Loci are said to be in linkage disequilibrium when the frequency of association of their different alleles is h ...
* Structural motif
References
* * {{Authority control Classical genetics Population genetics