In situ hybridization on:
[Wikipedia]
[Google]
[Amazon]
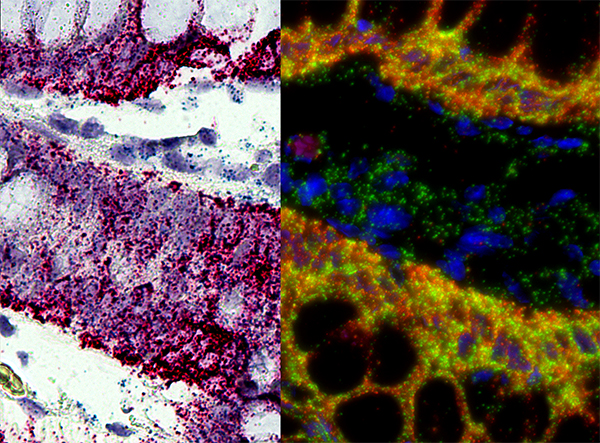 ''In situ'' hybridization (ISH) is a type of hybridization that uses a labeled
''In situ'' hybridization (ISH) is a type of hybridization that uses a labeled
Comprehensive and annotated in situ hybridization histochemistryRNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis
The Local Transcriptome in the Synaptic Neuropil Revealed by Deep Sequencing and High-Resolution Imaging
In Situ Hybridization of RNA and miRNA Probes to cells, CTCs, and tissuesWhole-Mount In Situ Hybridization of RNA Probes to Plant TissuesPreparation of Complex DNA Probe Sets for 3D FISH with up to Six Different FluorochromesTranscript In Situ Hybridization of Whole-Mount Embryos for Phenotype Analysis of RNAi-Treated Drosophila
* in-situ databases: *
*
Zebrafish gene expression
*
Mouse MGI GXD gene expression
*
eurexpress
Biochemistry detection methods Genetics techniques Laboratory techniques Biological techniques and tools
 ''In situ'' hybridization (ISH) is a type of hybridization that uses a labeled
''In situ'' hybridization (ISH) is a type of hybridization that uses a labeled complementary DNA
In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA (e.g., messenger RNA (mRNA) or microRNA (miRNA)) template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to express a spec ...
, RNA or modified nucleic acids strand (i.e., probe) to localize a specific DNA or RNA sequence in a portion or section of tissue (''in situ
''In situ'' (; often not italicized in English) is a Latin phrase that translates literally to "on site" or "in position." It can mean "locally", "on site", "on the premises", or "in place" to describe where an event takes place and is used in ...
'') or if the tissue is small enough (e.g., plant seeds, ''Drosophila
''Drosophila'' () is a genus of flies, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or (less frequently) pomace flies, vinegar flies, or wine flies, a reference to the characteristic of many speci ...
'' embryos), in the entire tissue (whole mount ISH), in cells, and in circulating tumor cells A circulating tumor cell (CTC) is a cell that has shed into the vasculature or lymphatics from a primary tumor and is carried around the body in the blood circulation. CTCs can extravasate and become ''seeds'' for the subsequent growth of additiona ...
(CTCs). This is distinct from immunohistochemistry, which usually localizes proteins in tissue sections.
In situ hybridization is used to reveal the location of specific nucleic acid sequences on chromosomes or in tissues, a crucial step for understanding the organization, regulation, and function of genes. The key techniques currently in use include ''in situ'' hybridization to mRNA with oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids ...
and RNA probes (both radio-labeled and hapten-labeled), analysis with light and electron microscopes, whole mount ''in situ'' hybridization, double detection of RNAs and RNA plus protein, and fluorescent ''in situ'' hybridization to detect chromosomal sequences. DNA ISH can be used to determine the structure of chromosomes. Fluorescent DNA ISH (FISH) can, for example, be used in medical diagnostics to assess chromosomal integrity. RNA ISH (RNA ''in situ'' hybridization) is used to measure and localize RNAs (mRNAs, lncRNAs, and miRNAs) within tissue sections, cells, whole mounts, and circulating tumor cells (CTCs). ''In situ'' hybridization was invented by American biologists Mary-Lou Pardue and Joseph G. Gall.
Challenges of in-situ hybridization
In situ hybridization is a powerful technique for identifying specific mRNA species within individual cells in tissue sections, providing insights into physiological processes and disease pathogenesis. However, in situ hybridization requires that many steps be taken with precise optimization for each tissue examined and for each probe used. In order to preserve the target mRNA within tissues, it is often required that crosslinking fixatives (such asformaldehyde
Formaldehyde ( , ) (systematic name methanal) is a naturally occurring organic compound with the formula and structure . The pure compound is a pungent, colourless gas that polymerises spontaneously into paraformaldehyde (refer to section ...
) be used.
In addition, in-situ hybridization on tissue sections require that tissue slices be very thin, usually 3 µm to 7 µm in thickness. Common methods of preparing tissue sections for in-situ hybridization processing include cutting specimens with a cryostat or a Compresstome tissue slicer. A cryostat
A cryostat (from ''cryo'' meaning cold and ''stat'' meaning stable) is a device used to maintain low cryogenic temperatures of samples or devices mounted within the cryostat. Low temperatures may be maintained within a cryostat by using various r ...
takes fresh or fixed tissue and immerses it into liquid nitrogen for flash freezing. Then tissue is embedded in freeze media called OCT and thin sections are cut. Obstacles include getting freeze artifacts on tissue that may interfere with proper mRNA staining. The Compresstome cuts tissue into thin slices without a freeze process; free-floating sections are cut after being embedded in agarose for stability. This method avoids freezing tissue and thus associated freeze artifacts. The process is permanent and irreversible once its complete.
Process
For hybridizationhistochemistry
Immunohistochemistry (IHC) is the most common application of immunostaining. It involves the process of selectively identifying antigens (proteins) in cells of a tissue section by exploiting the principle of antibodies binding specifically to ant ...
, sample cells and tissues are usually treated to fix the target transcripts in place and to increase access of the probe. As noted above, the probe is either a labeled complementary DNA
In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA (e.g., messenger RNA (mRNA) or microRNA (miRNA)) template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to express a spec ...
or, now most commonly, a complementary RNA (riboprobe A Riboprobe, abbreviation of RNA probe, is a segment of labelled RNA that can be used to detect a target mRNA or DNA during in situ hybridization. RNA probes can be produced by ''in vitro'' transcription of cloned DNA inserted in a suitable plasmi ...
). The probe hybridizes to the target sequence at elevated temperature, and then the excess probe is washed away (after prior hydrolysis using RNase in the case of unhybridized, excess RNA probe). Solution parameters such as temperature, salt, and/or detergent concentration can be manipulated to remove any non-identical interactions (i.e., only exact sequence matches will remain bound). Then, the probe that was labeled with either radio-, fluorescent- or antigen-labeled bases (e.g., digoxigenin
Digoxigenin (DIG) is a steroid found exclusively in the flowers and leaves of the plants '' Digitalis purpurea'', '' Digitalis orientalis'' and '' Digitalis lanata'' (foxgloves), where it is attached to sugars, to form the glycosides (e.g. Lana ...
) is localized and quantified in the tissue using either autoradiography, fluorescence microscopy
A fluorescence microscope is an optical microscope that uses fluorescence instead of, or in addition to, scattering, reflection, and attenuation or absorption, to study the properties of organic or inorganic substances. "Fluorescence microscop ...
, or immunohistochemistry, respectively. ISH can also use two or more probes, labeled with radioactivity or the other non-radioactive labels, to simultaneously detect two or more transcripts.
An alternative technology, branched DNA assay, can be used for RNA (mRNA, lncRNA, and miRNA ) ''in situ'' hybridization assays with single molecule sensitivity without the use of radioactivity. This approach (e.g., ViewRNA assays) can be used to visualize up to four targets in one assay, and it uses patented probe design and bDNA signal amplification to generate sensitive and specific signals. Samples (cells, tissues, and CTCs) are fixed, then treated to allow RNA target accessibility (RNA un-masking). Target-specific probes hybridize to each target RNA. Subsequent signal amplification is predicated on specific hybridization of adjacent probes (individual oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids ...
s ligosthat bind side by side on RNA targets). A typical target-specific probe will contain 40 oligonucleotides, resulting in 20 oligo pairs that bind side-by-side on the target for detection of mRNA and lncRNA, and 2 oligos or a single pair for miRNA detection. Signal amplification is achieved via a series of sequential hybridization steps. A pre-amplifier molecule hybridizes to each oligo pair on the target-specific RNA, then multiple amplifier molecules hybridize to each pre-amplifier. Next, multiple label probe oligonucleotides (conjugated to alkaline phosphatase or directly to fluorophores) hybridize to each amplifier molecule. A fully assembled signal amplification structure “Tree” has 400 binding sites for the label probes. When all target-specific probes bind to the target mRNA transcript, an 8,000 fold signal amplification occurs for that one transcript. Separate but compatible signal amplification systems enable the multiplex assays. The signal can be visualized using a fluorescence or brightfield microscope.
Basic steps for digoxigenin-labeled probes
# permeabilization of cells withproteinase K
In molecular biology Proteinase K (, ''protease K'', ''endopeptidase K'', ''Tritirachium alkaline proteinase'', ''Tritirachium album serine proteinase'', ''Tritirachium album proteinase K'') is a broad-spectrum serine protease. The enzyme was dis ...
to open cell membranes (around 25 minutes, not needed for tissue sections or some early-stage embryos)
# binding of mRNAs to marked RNA probe (usually overnight)
# antibody-phosphatase binding to RNA-probe (some hours)
# staining of antibody (e.g., with alkaline phosphatase)
The protocol takes around 2–3 days and takes some time to set up. Some companies sell robots to automate the process (e.g., Intavis InsituPro VSi). As a result, large-scale screenings have been conducted in laboratories on thousands of genes. The results can usually be accessed via websites (see external links).
See also
* Chromogenic ''in situ'' hybridization (CISH) * Fluorescence in situ hybridizationReferences
#Comprehensive and annotated in situ hybridization histochemistry
The Local Transcriptome in the Synaptic Neuropil Revealed by Deep Sequencing and High-Resolution Imaging
External links
* {{MeshName, In+Situ+HybridizationIn Situ Hybridization of RNA and miRNA Probes to cells, CTCs, and tissues
* in-situ databases: *
*
Zebrafish gene expression
*
Mouse MGI GXD gene expression
*
eurexpress
Biochemistry detection methods Genetics techniques Laboratory techniques Biological techniques and tools