uncompetitive inhibitor on:
[Wikipedia]
[Google]
[Amazon]
 Uncompetitive inhibition, also known as anti-competitive inhibition, takes place when an
Uncompetitive inhibition, also known as anti-competitive inhibition, takes place when an

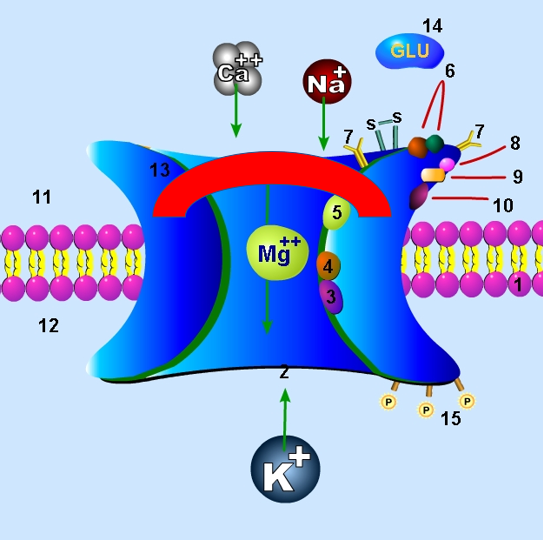 Uncompetitive inhibition can play roles in various other parts of the body as well. It is part of the mechanism by which NMDA (N-methyl-D-aspartate) glutamate receptors are inhibited in the brain, for example. Specifically, this type of inhibition impacts the granule cells that make up a layer of the cerebellum. These cells have the aforementioned NMDA receptors, and the activity of said receptors typically increases as ethanol is consumed. This often leads to withdrawal symptoms if said ethanol is removed. Various uncompetitive blockers act as antagonists at the receptors and modify the process, with one example being an inhibitor called
Uncompetitive inhibition can play roles in various other parts of the body as well. It is part of the mechanism by which NMDA (N-methyl-D-aspartate) glutamate receptors are inhibited in the brain, for example. Specifically, this type of inhibition impacts the granule cells that make up a layer of the cerebellum. These cells have the aforementioned NMDA receptors, and the activity of said receptors typically increases as ethanol is consumed. This often leads to withdrawal symptoms if said ethanol is removed. Various uncompetitive blockers act as antagonists at the receptors and modify the process, with one example being an inhibitor called
enzyme inhibitor
An enzyme inhibitor is a molecule that binds to an enzyme and blocks its activity. Enzymes are proteins that speed up chemical reactions necessary for life, in which substrate molecules are converted into products. An enzyme facilitates a sp ...
binds only to the complex formed between the enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products ...
and the substrate (the E-S complex). Uncompetitive inhibition typically occurs in reactions with two or more substrates or products.
While uncompetitive inhibition requires that an enzyme-substrate complex must be formed, non-competitive inhibition can occur with or without the substrate present.
Uncompetitive inhibition is distinguished from competitive inhibition by two observations: first uncompetitive inhibition cannot be reversed by increasing and second, as shown, the Lineweaver–Burk plot
In biochemistry, the Lineweaver–Burk plot (or double reciprocal plot) is a graphical representation of the Lineweaver–Burk equation of enzyme kinetics, described by Hans Lineweaver and Dean Burk in 1934. The Lineweaver–Burk plot for inhibit ...
yields parallel rather than intersecting lines. This behavior is found in the inhibition of acetylcholinesterase by tertiary amines (R3N). Such compounds bind to the enzyme in its various forms, but the acyl-intermediate-amine complex cannot break down into enzyme plus product.
Mechanism
As inhibitor binds, the amount of ES complex is reduced. This reduction in the effective concentration of the ES complex can be explained by the fact that having the inhibitor bound to the ES complex essentially converts it to ESI complex, which is considered a separate complex altogether. This reduction in ES complex decreases the maximum enzyme activity (Vmax), as it takes longer for the substrate or product to leave the active site. The reduction in Km - the substrate concentration at which the enzyme can operate at half of its maximal velocity, often used to approximate an enzyme's affinity for a substrate - can also be linked back to the decrease in ES complex. Le Chatelier's principle opposes this decrease and attempts to make up for the loss of ES, so more free enzyme is converted to the ES form, and the amount of ES increases overall. An increase in ES generally indicates that the enzyme has a high degree of affinity for its substrate. Km decreases as affinity for a substrate increases, though it is not a perfect predictor of affinity since it accounts for other factors as well; regardless, this increase in affinity will be accompanied by a decrease in Km. In general, uncompetitive inhibition works best when substrate concentration is high. An uncompetitive inhibitor need not resemble the substrate of the reaction it is inhibiting. At no concentration of substrate will the activity of the enzyme be higher when an uncompetitive inhibitor is present, but at low concentrations of substrate the enzyme activity difference will be negligible.Mathematical definition
The Lineweaver–Burk equation states that: : Where ''v'' is the initial reaction velocity, ''K''''m'' is the Michaelis–Menten constant, ''V''max is the maximum reaction velocity, and 'S''is theconcentration
In chemistry, concentration is the abundance of a constituent divided by the total volume of a mixture. Several types of mathematical description can be distinguished: '' mass concentration'', ''molar concentration'', '' number concentration'', ...
of the substrate.
The Lineweaver–Burk plot for an uncompetitive inhibitor produces a line parallel to the original enzyme-substrate plot, but with a higher y-intercept, due to the presence of an inhibition term :
:
Where 'I''is the concentration of the inhibitor and ''K''''i'' is an inhibition constant characteristic of the inhibitor.
The Michaelis-Menten equation is altered to:
:
where
: and
As described by the equation above, at high concentrations of substrate, ''V''''0'' approaches ''V''''max''/α'. Thus, an uncompetitive inhibitor lowers the measured ''V''''max''. Apparent ''K''''m'' also decreases, because required to reach one-half ''V''''max'' decreases by the factor α'. It is important to note that V''max'' and K''m'' decrease at the same rate as a result of the inhibitor. This is apparent when viewing a Lineweaver-Burk plot of uncompetitive enzyme inhibition: the ratio between V and K''m'' remains the same with or without an inhibitor present.
Implications and uses in biological systems
The unique traits of uncompetitive inhibition lead to a variety of implications for the inhibition's effects within biological and biochemical systems. Uncompetitive inhibition is present within biological systems in a number of ways. In fact, it often becomes clear that the traits of inhibition specific to uncompetitive inhibitors, such as their tendency to act at their best at high concentrations of substrate, are essential to some important bodily functions operating properly.Involvement in cancer mechanisms
Uncompetitive mechanisms are involved with certain types of cancer. Human alkaline phosphatases such as CGAP have been found to be over-expressed in certain types of cancers, and those phosphatases often operate via uncompetitive inhibition. It has also been found that a number of the genes that code for human alkaline phosphatases (TSAPs) are inhibited uncompetitively by amino acids such asleucine
Leucine (symbol Leu or L) is an essential amino acid that is used in the biosynthesis of proteins. Leucine is an α-amino acid, meaning it contains an α- amino group (which is in the protonated −NH3+ form under biological conditions), an α- ...
and phenylalanine. Studies of the involved amino acid residues have been undertaken in attempts to regulate alkaline phosphatase activity and learn more about said activity's relevance to cancer.
Additionally, uncompetitive inhibition works alongside TP53
p53, also known as Tumor protein P53, cellular tumor antigen p53 (UniProt name), or transformation-related protein 53 (TRP53) is a regulatory protein that is often mutated in human cancers. The p53 proteins (originally thought to be, and often ...
to help repress the activity of cancer cells and prevent tumorigenesis in certain forms of the illness, as it inhibits G6PD
Glucose-6-phosphate dehydrogenase (G6PD or G6PDH) () is a cytosolic enzyme that catalyzes the chemical reaction
: D-glucose 6-phosphate + NADP+ + H2O 6-phospho-D-glucono-1,5-lactone + NADPH + H+
This enzyme participates in the pentose phosp ...
(glucose-6-phosphate dehydrogenase, an enzyme primarily involved in certain metabolic pathways). One of the side roles G6PD is responsible for is helping to regulate is the control of reactive oxygen levels, as reactive oxygen species must be kept at appropriate levels to allow cells to survive. When G6PD's substrate concentration is high, uncompetitive inhibition of the enzyme becomes far more effective. As substrate concentration increases, the amount of ES complex increases as well, and with more ES complex to bind, uncompetitive inhibitors become far more active. This inhibition works such that the higher the concentration of substrate is in the system initially, the harder it is to reach the maximum velocity of the reaction. At low initial substrate concentrations, increasing the concentration of substrate is sometimes enough to entirely or even fully restore the enzyme's function, but as soon as initial concentration increases past a certain point, reaching the maximal enzyme velocity is all but impossible. This extreme sensitivity to substrate concentration within the cancer mechanism implicates uncompetitive inhibition rather than mixed inhibition, which displays similar traits but is often less sensitive to substrate concentration due to some inhibitor binding to free enzymes regardless of the substrate's presence. As such, the extreme strength of uncompetitive inhibitors at high substrate concentrations and the overall sensitivity to substrate amount indicates that only uncompetitive inhibition can make this type of process possible.
Importance in cell and organelle membranes
Although this form of inhibition is present in various diseases within biological systems, it does not necessarily only relate to pathologies. It can be involved in typical bodily functions. For example, active sites capable of uncompetitive inhibition appear to be present in membranes, as removing lipids from cell membranes and making active sites more accessible through conformational changes has been shown to invoke elements resembling the effects of uncompetitive inhibition (i.e. both KM and VMax decrease). In mitochondrial membrane lipids specifically, removing lipids decreases the alpha-helix content in mitochondria and leads to changes in ATPase resembling uncompetitive inhibition. This presence of uncompetitive enzymes in membranes has also been supported in a number of other studies. A type of protein called an Arf protein involved in regulating membrane activity was being studied, and it was found that an inhibitor called BFA trapped one of Arf's intermediates via uncompetitive inhibition. This made it clear that this type of inhibition exists within various types of cells and organelles as opposed to just in pathological cells. In fact, BFA was found to relate to the activity of the Golgi apparatus and its role in regulating movement across the cell membrane.Presence in the cerebellar granule layer
 Uncompetitive inhibition can play roles in various other parts of the body as well. It is part of the mechanism by which NMDA (N-methyl-D-aspartate) glutamate receptors are inhibited in the brain, for example. Specifically, this type of inhibition impacts the granule cells that make up a layer of the cerebellum. These cells have the aforementioned NMDA receptors, and the activity of said receptors typically increases as ethanol is consumed. This often leads to withdrawal symptoms if said ethanol is removed. Various uncompetitive blockers act as antagonists at the receptors and modify the process, with one example being an inhibitor called
Uncompetitive inhibition can play roles in various other parts of the body as well. It is part of the mechanism by which NMDA (N-methyl-D-aspartate) glutamate receptors are inhibited in the brain, for example. Specifically, this type of inhibition impacts the granule cells that make up a layer of the cerebellum. These cells have the aforementioned NMDA receptors, and the activity of said receptors typically increases as ethanol is consumed. This often leads to withdrawal symptoms if said ethanol is removed. Various uncompetitive blockers act as antagonists at the receptors and modify the process, with one example being an inhibitor called memantine
Memantine is a medication used to slow the progression of moderate-to-severe Alzheimer's disease. It is taken by mouth.
Common side effects include headache, constipation, sleepiness, and dizziness. Severe side effects may include blood clots ...
. In fact, in similar cases (involving over-expression of NMDA, though not necessarily via ethanol), it has been shown that uncompetitive inhibition helps in nullifying the over-expression due to its particular properties. Since uncompetitive inhibitors block high concentrations of substrates very efficiently, their traits alongside the innate characteristics of the receptors themselves lead to very effective blocking of NMDA channels when they are excessively open due to massive amounts of NMDA agonists.
Derivation of Uncompetitive Michaelis–Menten Equation
Uncompetitive inhibition can be described using the following set of reaction equations: The reaction rate of the first rule defined above where and bind to form the complex Sand Scan separate to reform the products can be written as: