internal transcribed spacer on:
[Wikipedia]
[Google]
[Amazon]
Internal transcribed spacer (ITS) is the spacer DNA situated between the small-subunit ribosomal RNA (rRNA) and large-subunit rRNA genes in the
 In bacteria and archaea, the ITS occurs in one to several copies, as do the flanking 16S and
In bacteria and archaea, the ITS occurs in one to several copies, as do the flanking 16S and
University of Washington Laboratory Medicine: Molecular Diagnosis , Yeast Sequencing
ITSone DB
ITS2 database
(Schultz et al.) {{DEFAULTSORT:Internal Transcribed Spacer DNA Phylogenetics
chromosome
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins ar ...
or the corresponding transcribed region in the polycistronic
A cistron is an alternative term for " gene". The word cistron is used to emphasize that genes exhibit a specific behavior in a cis-trans test; distinct positions (or loci) within a genome are cistronic.
History
The words ''cistron'' and ''ge ...
rRNA precursor transcript.
ITS across life domains
In bacteria and archaea, there is a single ITS, located between the 16S and23S
The 23S rRNA is a 2,904 nucleotide long (in '' E. coli'') component of the large subunit (50S) of the bacterial/archean ribosome and makes up the peptidyl transferase center (PTC). The 23S rRNA is divided into six secondary structural domains ...
rRNA genes. Conversely, there are two ITSs in eukaryotes: ITS1 is located between 18S and 5.8S rRNA genes, while ITS2 is between 5.8S and 28S
28S ribosomal RNA is the structural ribosomal RNA (rRNA) for the large subunit (LSU) of eukaryotic cytoplasmic ribosomes, and thus one of the basic components of all eukaryotic cells. It has a size of 25S in plants and 28S in mammals, hence th ...
(in opisthokonts, or 25S in plants) rRNA genes. ITS1 corresponds to the ITS in bacteria and archaea, while ITS2 originated as an insertion that interrupted the ancestral 23S rRNA gene.
Organization
 In bacteria and archaea, the ITS occurs in one to several copies, as do the flanking 16S and
In bacteria and archaea, the ITS occurs in one to several copies, as do the flanking 16S and 23S
The 23S rRNA is a 2,904 nucleotide long (in '' E. coli'') component of the large subunit (50S) of the bacterial/archean ribosome and makes up the peptidyl transferase center (PTC). The 23S rRNA is divided into six secondary structural domains ...
genes. When there are multiple copies, these do not occur adjacent to one another. Rather, they occur in discrete locations in the circular chromosome. It is not uncommon in bacteria to carry tRNA genes in the ITS.
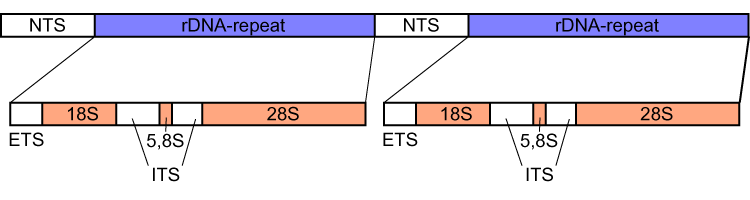
In eukaryotes, genes encoding ribosomal RNA and spacers occur in tandem repeats that are thousands of copies long, each separated by regions of non-transcribed DNA termed '' intergenic spacer'' (IGS) or ''non-transcribed spacer'' (NTS).
Each eukaryotic ribosomal cluster contains the 5' external transcribed spacer
External transcribed spacer (ETS) refers to a piece of non-functional RNA, closely related to the internal transcribed spacer, which is situated outside structural ribosomal RNAs (rRNA) on a common precursor transcript. ETS sequences characteristi ...
(5' ETS), the 18S rRNA gene, the ITS1
Internal transcribed spacer (ITS) is the spacer DNA situated between the small-subunit ribosomal RNA (rRNA) and large-subunit rRNA genes in the chromosome or the corresponding transcribed region in the polycistronic rRNA precursor transcript.
...
, the 5.8S rRNA gene, the ITS2
Internal transcribed spacer (ITS) is the spacer DNA situated between the small-subunit ribosomal RNA (rRNA) and large-subunit rRNA genes in the chromosome or the corresponding transcribed region in the polycistronic rRNA precursor transcript.
...
, the 26S or 28S rRNA gene, and finally the 3' ETS.
During rRNA maturation, ETS and ITS pieces are excised. As non-functional by-products of this maturation, they are rapidly degraded.
Use in phylogenetic inference
Sequence comparison of the eukaryotic ITS regions is widely used in taxonomy and molecular phylogeny because of several favorable properties: * It is routinely amplified thanks to its small size associated to the availability of highly conserved flanking sequences. * It is easy to detect even from small quantities of DNA due to the high copy number of the rRNA clusters. * It undergoes rapid concerted evolution via unequal crossing-over and gene conversion. This promotes intra-genomic homogeneity of the repeat units, although high-throughput sequencing showed the occurrence of frequent variations within plant species. * It has a high degree of variation even between closely related species. This can be explained by the relatively low evolutionary pressure acting on such non-coding spacer sequences. For example, ITS markers have proven especially useful for elucidating phylogenetic relationships among the following taxa. ITS2 is known to be more conserved than ITS1 is. All ITS2 sequences share a common core of secondary structure, while ITS1 structures are only conserved in much smaller taxonomic units. Regardless of the scope of conservation, structure-assisted comparison can provide higher resolution and robustness.Mycological barcoding
The ITS region is the most widely sequenced DNA region in molecular ecology offungi
A fungus ( : fungi or funguses) is any member of the group of eukaryotic organisms that includes microorganisms such as yeasts and molds, as well as the more familiar mushrooms. These organisms are classified as a kingdom, separately fr ...
and has been recommended as the universal fungal barcode sequence. It has typically been most useful for molecular systematics at the species to genus level, and even within species (e.g., to identify geographic races). Because of its higher degree of variation than other genic regions of rDNA (for example, small- and large-subunit rRNA), variation among individual rDNA repeats can sometimes be observed within both the ITS and IGS regions. In addition to the universal ITS1+ITS4 primers used by many labs, several taxon-specific primers have been described that allow selective amplification of fungal sequences (e.g., see Gardes & Bruns 1993 paper describing amplification of basidiomycete
Basidiomycota () is one of two large divisions that, together with the Ascomycota, constitute the subkingdom Dikarya (often referred to as the "higher fungi") within the kingdom Fungi. Members are known as basidiomycetes. More specifically, Bas ...
ITS sequences from mycorrhiza samples). Despite shotgun sequencing
In genetics, shotgun sequencing is a method used for sequencing random DNA strands. It is named by analogy with the rapidly expanding, quasi-random shot grouping of a shotgun.
The chain-termination method of DNA sequencing ("Sanger sequencing ...
methods becoming increasingly utilized in microbial sequencing, the low biomass of fungi in clinical samples make the ITS region amplification an area of ongoing research.
References
External links
University of Washington Laboratory Medicine: Molecular Diagnosis , Yeast Sequencing
ITSone DB
ITS2 database
(Schultz et al.) {{DEFAULTSORT:Internal Transcribed Spacer DNA Phylogenetics