XDNA on:
[Wikipedia]
[Google]
[Amazon]
xDNA (also known as expanded DNA or benzo-homologated DNA) is a size-expanded nucleotide system synthesized from the fusion of a benzene ring and one of the four natural bases: adenine, guanine, cytosine, and thymine. This size expansion produces an 8 letter alphabet which has a larger information density by a factor of 2n compared to natural DNA's (often referred to as B-DNA in literature) 4 letter alphabet. As with normal base-pairing, A pairs with xT, C pairs with xG, G pairs with xC, and T pairs with xA. The double helix is thus 2.4 Ã… wider than a natural double helix. While similar in structure to B-DNA, xDNA has unique absorption, fluorescence, and stacking properties.
Initially synthesized as an enzyme probe by Nelson J. Leonard's group, benzo-homologated adenine was the first base synthesized. Later,
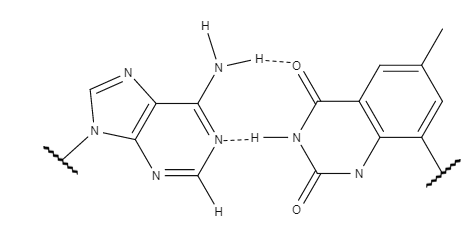 The success and implications of xDNA prompted research to examine other factors which could alter B-DNA's chemical properties and create a new system for information storage with broader applications. yDNA also uses a benzene ring, similar to xDNA, with the only difference being the site of addition of the aromatic ring. The location of the benzene ring changes the preferred structure of the expanded helix. The altered conformation makes yDNA more similar to B-DNA in its orientation by changing the interstrand
The success and implications of xDNA prompted research to examine other factors which could alter B-DNA's chemical properties and create a new system for information storage with broader applications. yDNA also uses a benzene ring, similar to xDNA, with the only difference being the site of addition of the aromatic ring. The location of the benzene ring changes the preferred structure of the expanded helix. The altered conformation makes yDNA more similar to B-DNA in its orientation by changing the interstrand 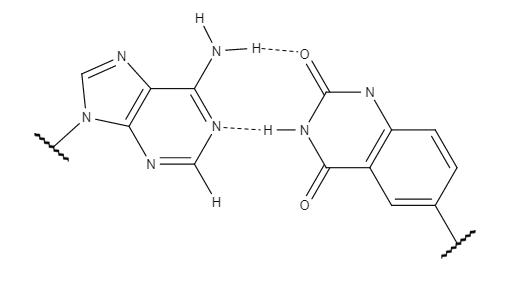 While xDNA and yDNA are quite similar in most properties, including their increased stacking interactions, yDNA shows superior mismatch recognition. y-pyrimidines display slightly stronger stacking interactions than x-pyrimidines as a result of the distance between the two
While xDNA and yDNA are quite similar in most properties, including their increased stacking interactions, yDNA shows superior mismatch recognition. y-pyrimidines display slightly stronger stacking interactions than x-pyrimidines as a result of the distance between the two
 Doubly-expanded (or ''naphtho-homologated'') nucleobases incorporate a
Doubly-expanded (or ''naphtho-homologated'') nucleobases incorporate a
Eric T. Kool
Eric T. Kool is an American chemist, focusing in chemistry of RNA and DNA; probe design and imaging; synthetic biology, currently the George A. and Hilda M. Daubert Professor in Chemistry at Stanford University and is an Elected Fellow at the Ameri ...
's group finished synthesizing the remaining three expanded bases, eventually followed by yDNA ("wide" DNA), another benzo-homologated nucleotide system, and naphtho-homologated xxDNA and yyDNA. xDNA is more stable when compared to regular DNA when subjected to higher temperature, and while entire strands of xDNA, yDNA, xxDNA and yyDNA exist, they are currently difficult to synthesize and maintain. Experiments with xDNA provide new insight into the behavior of natural B-DNA. The extended bases xA, xC, xG, and xT are naturally fluorescent, and single strands composed of only extended bases can recognize and bind to single strands of natural DNA, making them useful tools for studying biological systems. xDNA is most commonly formed with base pairs between a natural and expanded nucleobase, however x-nucleobases can also be paired together. Current research supports xDNA as a viable genetic encoding system in the near future.
Origins
The first nucleotide to be expanded was the purine adenine. Nelson J. Leonard and colleagues synthesized this original x-nucleotide, which was referred to as "expanded adenine". xA was used as a probe in the investigation ofactive site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate (binding site) a ...
s of ATP
ATP may refer to:
Companies and organizations
* Association of Tennis Professionals, men's professional tennis governing body
* American Technical Publishers, employee-owned publishing company
* ', a Danish pension
* Armenia Tree Project, non ...
-dependent enzymes, more specifically what modifications the substrate
Substrate may refer to:
Physical layers
*Substrate (biology), the natural environment in which an organism lives, or the surface or medium on which an organism grows or is attached
** Substrate (locomotion), the surface over which an organism lo ...
could take while still being functional. Almost two decades later, the other three bases were successfully expanded and later integrated into a double helix by Eric T. Kool
Eric T. Kool is an American chemist, focusing in chemistry of RNA and DNA; probe design and imaging; synthetic biology, currently the George A. and Hilda M. Daubert Professor in Chemistry at Stanford University and is an Elected Fellow at the Ameri ...
and colleagues. Their goal was to create a synthetic genetic system which mimics and surpasses the functions of the natural genetic system, and to broaden the applications of DNA both in living cells and in experimental biochemistry. Once the expanded base set was created, the goal shifted to identifying or developing faithful replication enzymes and further optimizing the expanded DNA alphabet.
Synthesis
In benzo-homologated purines (xA and xG), the benzene ring is bound to the nitrogenous base through nitrogen-carbon (N-C) bonds. Benzo-homologated pyrimidines are formed through carbon-carbon (C-C) bonds between the base and the benzene. Thus far, x-nucleobases have been added to strands of DNA using phosphoramidite derivatives, as traditional polymerases have been unsuccessful in synthesizing strands of xDNA. X-nucleotides are poor candidates as substrates for B-DNA polymerases as their size interferes with binding at thecatalytic domain
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate ( binding si ...
. Attempts at using template-independent enzymes have been successful as they have a reduced geometric constraint for substrates. Terminal deoxynucleotidyl transferase (TdT) has been used previously to synthesize strands of bases which have been bound to fluorophores. Using TdT
Terminal deoxynucleotidyl transferase (TdT), also known as DNA nucleotidylexotransferase (DNTT) or terminal transferase, is a specialized DNA polymerase expressed in immature, pre-B, pre-T lymphoid cells, and acute lymphoblastic leukemia/lymphom ...
, up to 30 monomers can be combined to form a double-helix of xDNA, however this oligomer
In chemistry and biochemistry, an oligomer () is a molecule that consists of a few repeating units which could be derived, actually or conceptually, from smaller molecules, monomers.Quote: ''Oligomer molecule: A molecule of intermediate relativ ...
ic xDNA appears to inhibit its own extension beyond this length due to the overwhelming hydrogen bonding. In order to minimize inhibition, xDNA can be hybridized into a regular helix.
Replication
For xDNA to be used as a substitute structure for information storage, it requires a reliable replication mechanism. Research into xDNA replication using aKlenow fragment
The Klenow fragment is a large protein fragment produced when DNA polymerase I from '' E. coli'' is enzymatically cleaved by the protease subtilisin. First reported in 1970, it retains the 5' → 3' polymerase activity and the 3’ → 5’ ex ...
from DNA polymerase I shows that a natural base partner is selectively added in instances of single-nucleotide insertion. However, DNA polymerase IV
DNA polymerase IV is a prokaryotic polymerase that is involved in mutagenesis and is encoded by the ''dinB'' gene. It exhibits no 3′→5′ exonuclease (proofreading) activity and hence is error prone. In ''E. coli'', DNA polymerase IV (Pol 4) ...
(Dpo4) has been able to successfully use xDNA for these types of insertions with high fidelity, making it a promising candidate for future research in extending replicates of xDNA. xDNA's mismatch sensitivity is similar to that of B-DNA.
Structure
Similar to natural bases, x-nucleotides selectively assemble into a duplex-structure resembling B-DNA. xDNA was originally synthesized by incorporating a benzene ring into the nitrogenous base. However, other expanded bases have been able to incorporate thiophene and benzo hiophene as well. xDNA and yDNA use benzene rings to widen the bases and are thus termed "benzo-homologated". Another form of expanded nucleobases known as yyDNA incorporatenaphthalene
Naphthalene is an organic compound with formula . It is the simplest polycyclic aromatic hydrocarbon, and is a white crystalline solid with a characteristic odor that is detectable at concentrations as low as 0.08 ppm by mass. As an aromati ...
into the base and are "naptho-homologated". xDNA has a rise of 3.2 Å and a twist of 32°, significantly smaller than B-DNA, which has a rise of 3.3 Å and a twist of 34.2° xDNA nucleotides can occur on both strands—either alone (known as "doubly expanded DNA") or mixed with natural bases—or exclusively on one strand or the other. Similar to B-DNA, xDNA can recognize and bind complementary single-stranded DNA or RNA
Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid ( DNA) are nucleic acids. Along with lipids, proteins, and carbohydra ...
sequences.
Duplexes formed from xDNA are similar to natural duplexes aside from the distance between the two sugar-phosphate backbones. xDNA helices have a greater number of base pairs per turn of the helix as a result of a reduced distance between neighbour nucleotides. NMR spectra report that xDNA helices are anti-parallel, right-handed and take an ''anti'' conformation around the glycosidic bond, with a C2'-endo sugar pucker. Helices created from xDNA are more likely to take a B-helix over an A-helix conformation, and have an increased major groove width by 6.5 Ã… (where the backbones are farthest apart) and decreased minor groove width by 5.5 Ã… (where the backbones are closest together) compared to B-DNA. Altering groove width affects the xDNA's ability to associate with DNA-binding proteins, but as long as the expanded nucleotides are exclusive to one strand, recognition sites are sufficiently similar to B-DNA to allow bonding of transcription factors and small polyamide molecules. Mixed helices present the possibility of recognizing the four expanded bases using other DNA-binding molecules.
Properties
Expanded nucleotides and their oligomeric helices share many properties with their natural B-DNA counterparts, including their pairing preference: A with T, C with G. The various differences in chemical properties between xDNA and B-DNA support the hypothesis that the benzene ring which expands x-nucleobases is not, in fact, chemically inert. xDNA is more hydrophobic than B-DNA, and also has a smaller HOMO-LUMO gap (distance between the highest occupied molecular orbital and lowest unoccupied molecular orbital) as a result of modified saturation. xDNA has higher melting temperatures than B-DNA (a mixed decamer of xA and T has a melting temperature of 55.6 °C, 34.3 °C higher than the same decamer of A and T), and exhibits an "all-or-nothing" melting behaviour.Conformation
Under lab conditions, xDNA orients itself in the ''syn'' conformation. This unfortunately does not expose the binding face of the xDNA nucleotides to face the neighbouring strand for binding, meaning that extra measures must be applied to alter the conformation of xDNA before attempting to form helices. However, the ''anti'' and ''syn'' orientations are practically identical energetically in expanded bases. This conformational preference is seen primarily inpyrimidine
Pyrimidine (; ) is an aromatic, heterocyclic, organic compound similar to pyridine (). One of the three diazines (six-membered heterocyclics with two nitrogen atoms in the ring), it has nitrogen atoms at positions 1 and 3 in the ring. The other ...
s, and purines display minimal preference for orientation.
Enhanced stacking
Stacking of the nucleotides in a double helix is a major determinant of the helix's stability. With the addedsurface area
The surface area of a solid object is a measure of the total area that the surface of the object occupies. The mathematical definition of surface area in the presence of curved surfaces is considerably more involved than the definition of arc ...
and hydrogen available for bonding, stacking potential for the nucleobases increases with the addition of a benzene spacer. By increasing the separation between the nitrogenous bases and either sugar-phosphate backbone, the helix's stacking energy is less variable and therefore more stable. The energies for natural nucleobase pairs vary from 18 to 52 kJ/mol. This variance is only 14–40 kJ/mol for xDNA.
Due to an increased overlap between and expanded strand of DNA and its neighbouring strand, there are greater interstrand interactions in expanded and mixed helices, resulting in a significant increase in the helix's stability. xDNA has enhanced stacking abilities resultant from changes in inter- and intrastrand hydrogen bond
In chemistry, a hydrogen bond (or H-bond) is a primarily electrostatic force of attraction between a hydrogen (H) atom which is covalently bound to a more electronegative "donor" atom or group (Dn), and another electronegative atom bearing a ...
ing that arise from the addition of a benzene spacer, but expanding the bases does not alter hydrogen's contribution to the stability of the duplex. These stacking abilities are exploited by helices consisting of both xDNA and B-DNA in order to optimize the strength of the helix. Increased stacking is seen most prominently in strands consisting only of A and xA and T and xT, as T-xA has stronger stacking interactions than T- A.
The energy resultant from pyrimidine
Pyrimidine (; ) is an aromatic, heterocyclic, organic compound similar to pyridine (). One of the three diazines (six-membered heterocyclics with two nitrogen atoms in the ring), it has nitrogen atoms at positions 1 and 3 in the ring. The other ...
s ranges from 30 to 49 kJ/mol. The range for purines is between 40-58kJ/mol. By replacing one nucleotide in a double-helix with an expanded nucleotide, the strength of the stacking interactions increases by 50%. Expanding both nucleotides results in a 90% increase in stacking strength. While xG has an overall negative effect on the binding strength of the helix, the other three expanded bases outweigh this with their positive effects. The change in energy caused by expanding the bases is mostly dependent on the rotation of the bond about the nucleobases' centers of mass
In physics, the center of mass of a distribution of mass in space (sometimes referred to as the balance point) is the unique point where the weighted relative position of the distributed mass sums to zero. This is the point to which a force may ...
, and center of mass stacking interactions improve the stacking potential of the helix. Because the size-expanded bases widen the helix, it is more thermally stable with a higher melting temperature.
Absorption
The addition of a benzene spacer in x- nucleobases affects the bases' optical absorption spectra. Time-dependent density functional theory (TDDFT) applied to xDNA revealed that the benzene component of the highest occupied molecular orbitals ( HOMO) in the x-bases pins the absorption onset at an earlier point than natural bases. Another unusual feature of xDNA absorption spectra is the red-shifted excimers of xA in the low range. In terms of stacking fingerprints, there is a more pronounced hypochromicity seen in consecutive xA- Tbase pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
s.
Implications of xDNA's altered absorption include applications in nanoelectronic technology and nanobiotechnology
Nanobiotechnology, bionanotechnology, and nanobiology are terms that refer to the intersection of nanotechnology and biology. Given that the subject is one that has only emerged very recently, bionanotechnology and nanobiotechnology serve as blan ...
. The reduced spacing between x-nucleotides makes the helix stiffer, thus it is not as easily affected by substrate
Substrate may refer to:
Physical layers
*Substrate (biology), the natural environment in which an organism lives, or the surface or medium on which an organism grows or is attached
** Substrate (locomotion), the surface over which an organism lo ...
, electrode, and functional nanoparticle forces. Other alterations to natural nucleotides resulting in different absorption spectra will broaden these applications in the future.
Fluorescence
One unique property of xDNA is its inherent fluorescence. Natural bases can be bound directly to fluorophores for use in microarrays, ''in situ'' hybridization, andpolymorphism
Polymorphism, polymorphic, polymorph, polymorphous, or polymorphy may refer to:
Computing
* Polymorphism (computer science), the ability in programming to present the same programming interface for differing underlying forms
* Ad hoc polymorphis ...
analysis. However, these fluorescent natural bases often fail as a result of self-quenching, which diminishes their fluorescent intensity and reduces their applicability as visual DNA tags. The pi interactions between the rings in x-nucleobases result in an inherent fluorescence in the violet-blue range, with a Stokes shift
__NOTOC__
Stokes shift is the difference (in energy, wavenumber or frequency units) between positions of the band maxima of the absorption and emission spectra (fluorescence and Raman being two examples) of the same electronic transition. It ...
between 50 and 80 nm. They also have a quantum yield in the range of 0.3–0.6. xC has the greatest fluorescent emission.
Other expanded bases
After the creation of and successful research surrounding xDNA, more forms of expanded nucleotides were investigated. yDNA is a second, similar system of nucleotides which uses a benzene ring to expand the four natural bases. xxDNA and yyDNA usenaphthalene
Naphthalene is an organic compound with formula . It is the simplest polycyclic aromatic hydrocarbon, and is a white crystalline solid with a characteristic odor that is detectable at concentrations as low as 0.08 ppm by mass. As an aromati ...
, a polycyclic molecule consisting of two hydrocarbon rings. The two rings expand the base even wider, further altering its chemical properties.
yDNA
 The success and implications of xDNA prompted research to examine other factors which could alter B-DNA's chemical properties and create a new system for information storage with broader applications. yDNA also uses a benzene ring, similar to xDNA, with the only difference being the site of addition of the aromatic ring. The location of the benzene ring changes the preferred structure of the expanded helix. The altered conformation makes yDNA more similar to B-DNA in its orientation by changing the interstrand
The success and implications of xDNA prompted research to examine other factors which could alter B-DNA's chemical properties and create a new system for information storage with broader applications. yDNA also uses a benzene ring, similar to xDNA, with the only difference being the site of addition of the aromatic ring. The location of the benzene ring changes the preferred structure of the expanded helix. The altered conformation makes yDNA more similar to B-DNA in its orientation by changing the interstrand hydrogen bond
In chemistry, a hydrogen bond (or H-bond) is a primarily electrostatic force of attraction between a hydrogen (H) atom which is covalently bound to a more electronegative "donor" atom or group (Dn), and another electronegative atom bearing a ...
s. Stability is highly dependent on the bases' rotation about the link between the base and the sugar of the backbone. yDNA's altered preference for this orientation makes it more stable overall than xDNA. The location of the benzene spacer also affects the bases' groove geometry, altering neighbour interactions. The base pairs between y-nucleotides and natural nucleotides is planar, rather than slightly twisted as with xDNA. This decreases the rise of the helix even further than achieved by xDNA.
 While xDNA and yDNA are quite similar in most properties, including their increased stacking interactions, yDNA shows superior mismatch recognition. y-pyrimidines display slightly stronger stacking interactions than x-pyrimidines as a result of the distance between the two
While xDNA and yDNA are quite similar in most properties, including their increased stacking interactions, yDNA shows superior mismatch recognition. y-pyrimidines display slightly stronger stacking interactions than x-pyrimidines as a result of the distance between the two anomer
In carbohydrate chemistry, a pair of anomers () is a pair of near-identical stereoisomers that differ at only the anomeric carbon, the carbon that bears the aldehyde or ketone functional group in the sugar's open-chain form. However, in order fo ...
ic carbons, which is slightly larger in yDNA. xDNA still has stronger stacking interactions in model helices, but adding either x- or y-pyrimidines to a natural double helix strengthens the intra- and interstrand interactions, increasing overall helix stability. In the end, which of the two has the strongest overall stacking interactions is dependent on the sequence; xT and yT bind A with similar strength, but the stacking energy of yC bound to G is stronger than xC by 4kJ/mol. yDNA and other expanded bases are part of a very young field which is highly understudied. Research suggest that the ideal conformation still remains to be discovered, but knowing that the benzene location affects the orientation and structure of expanded nucleobases adds information to their future design.
yyDNA and xxDNA
 Doubly-expanded (or ''naphtho-homologated'') nucleobases incorporate a
Doubly-expanded (or ''naphtho-homologated'') nucleobases incorporate a naphthalene
Naphthalene is an organic compound with formula . It is the simplest polycyclic aromatic hydrocarbon, and is a white crystalline solid with a characteristic odor that is detectable at concentrations as low as 0.08 ppm by mass. As an aromati ...
spacer instead of a benzene ring, widening the base twice as much with its two-ringed structure. These structures (known as xxDNA and yyDNA) are 4.8 Ã… wider than natural bases and were once again created as a result of Leonard's research on expanded adenine in ATP
ATP may refer to:
Companies and organizations
* Association of Tennis Professionals, men's professional tennis governing body
* American Technical Publishers, employee-owned publishing company
* ', a Danish pension
* Armenia Tree Project, non ...
-dependent enzymes in 1984. No literature was published on these doubly-expanded bases for nearly three decades until 2013 when the first xxG was produced by Sharma, Lait, and Wetmore and incorporated along with xxA into a natural helix. Although very little research has been performed on xxDNA, xx- purine neighbours have already been shown to increase intrastrand stacking energy by up to 119% (as opposed to 62% in x-purines). xx- purine and pyrimidine
Pyrimidine (; ) is an aromatic, heterocyclic, organic compound similar to pyridine (). One of the three diazines (six-membered heterocyclics with two nitrogen atoms in the ring), it has nitrogen atoms at positions 1 and 3 in the ring. The other ...
interactions show an overall decrease in stacking energies, but the overall stability of duplexes including pyrimidines and xx-purines increases by 22%, more than twofold that of pyrimidines and x-purines.
Uses
xDNA has many applications in chemical and biological research, including expanding upon applications of natural DNA, such as scaffolding. In order to create self-assembling nanostructures, a scaffold is needed as a sort oftrellis
Trellis may refer to:
Structures
* Trellis (architecture), an architectural structure often used to support plants (especially vineyards)
* Trellis drainage pattern, a drainage system
Technology
* Trellis (graph), a special kind of graph used ...
to support the growth. DNA has been used as a means to this end in the past, but expanded scaffolds make larger scaffolds for more complex self-assembly an option. xDNA's electrical conduction properties also make it a prime candidate as a molecular wire
Molecular wires (or sometimes called molecular nanowires) are molecular chains that conduct electric current. They are the proposed building blocks for molecular
electronic devices. Their typical diameters are less than three nanometers, while th ...
, as its π-π interactions help it efficiently conduct electricity. Its 8-letter alphabet ( A, T, C, G, xA, xT, xC, xG) gives it the potential to store 2n times increase in storage density, where ''n'' represents the number of letters in a sequence. For example, combining 6 nucleotides of with B-DNA yields 4096 possible sequences, whereas a combination of the same number of nucleotides created with xDNA yields 262,144 possible sequences. Additionally, xDNA can be used as a fluorescent probe at enzyme active site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate (binding site) a ...
s, as was its original application by Leonard et al.
xDNA has also been applied to the study of protein-DNA interactions. Due to xDNA's natural fluorescing
Fluorescence is the emission of light by a substance that has absorbed light or other electromagnetic radiation. It is a form of luminescence. In most cases, the emitted light has a longer wavelength, and therefore a lower photon energy, tha ...
properties, it can easily be visualized in both lab and living conditions. xDNA is becoming more easy to create and oligomer
In chemistry and biochemistry, an oligomer () is a molecule that consists of a few repeating units which could be derived, actually or conceptually, from smaller molecules, monomers.Quote: ''Oligomer molecule: A molecule of intermediate relativ ...
ize, and its high-affinity binding to complementary
A complement is something that completes something else.
Complement may refer specifically to:
The arts
* Complement (music), an interval that, when added to another, spans an octave
** Aggregate complementation, the separation of pitch-class ...
DNA and RNA
Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid ( DNA) are nucleic acids. Along with lipids, proteins, and carbohydra ...
sequences means that it can not only help locate these sequences floating around in the cell, but also when they are already interacting with other structures within the cell. xDNA also has potential applications in assays that employ TdT
Terminal deoxynucleotidyl transferase (TdT), also known as DNA nucleotidylexotransferase (DNTT) or terminal transferase, is a specialized DNA polymerase expressed in immature, pre-B, pre-T lymphoid cells, and acute lymphoblastic leukemia/lymphom ...
as it may improve reporters, and can be used as an affinity tag for interstrand bonding.
See also
* DNA *RNA
Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid ( DNA) are nucleic acids. Along with lipids, proteins, and carbohydra ...
* DNA sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. Th ...
* Genetic engineering
Genetic engineering, also called genetic modification or genetic manipulation, is the modification and manipulation of an organism's genes using technology. It is a set of technologies used to change the genetic makeup of cells, including t ...
* Nanobiotechnology
Nanobiotechnology, bionanotechnology, and nanobiology are terms that refer to the intersection of nanotechnology and biology. Given that the subject is one that has only emerged very recently, bionanotechnology and nanobiotechnology serve as blan ...
* Nucleobase
* Hachimoji DNA
* Artificially Expanded Genetic Information System (AEGIS)
References
{{Reflist Biochemistry methods DNA Genetics techniques Molecular biology