TET Enzymes on:
[Wikipedia]
[Google]
[Amazon]
The TET enzymes are a family of ten-eleven translocation (TET) methylcytosine dioxygenases. They are instrumental in  Demethylation by TET enzymes (see second Figure), can alter the regulation of transcription. The TET enzymes catalyze the
Demethylation by TET enzymes (see second Figure), can alter the regulation of transcription. The TET enzymes catalyze the  TET enzymes have central roles in
TET enzymes have central roles in
 The three mammalian
The three mammalian
 The mouse
The mouse
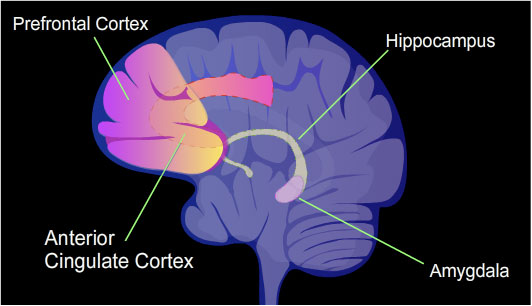 Learning and memory have levels of permanence, differing from other mental processes such as thought, language, and consciousness, which are temporary in nature. Learning and memory can be either accumulated slowly (multiplication tables) or rapidly (touching a hot stove), but once attained, can be recalled into conscious use for a long time. Rats subjected to one instance of contextual fear conditioning create an especially strong long-term memory. At 24 hours after training, 9.17% of the genes in the genomes of rat hippocampus neurons were found to be differentially methylated. This included more than 2,000 differentially methylated genes at 24 hours after training, with over 500 genes being demethylated. Similar results to that in the rat hippocampus were also obtained in mice with contextual fear conditioning.
The hippocampus region of the brain is where contextual fear memories are first stored (see Figure), but this storage is transient and does not remain in the hippocampus. In rats contextual fear conditioning is abolished when the hippocampus is subjected to hippocampectomy just one day after conditioning, but rats retain a considerable amount of contextual fear when hippocampectomy is delayed by four weeks. In mice, examined at 4 weeks after conditioning, the hippocampus methylations and demethylations were reversed (the hippocampus is needed to form memories but memories are not stored there) while substantial differential CpG methylation and demethylation occurred in cortical neurons during memory maintenance. There were 1,223 differentially methylated genes in the anterior cingulate cortex (see Figure) of mice four weeks after contextual fear conditioning. Thus, while there were many methylations in the hippocampus shortly after memory was formed, all these hippocampus methylations were demethylated as soon as four weeks later.
Li et al. reported one example of the relationship between expression of a TET protein, demethylation and memory while using extinction training. Extinction training is the disappearance of a previously learned behavior when the behavior is not reinforced.
A comparison between infralimbic prefrontal cortex (ILPFC) neuron samples derived from mice trained to fear an auditory cue and extinction-trained mice revealed dramatic experience-dependent genome-wide differences in the accumulation of 5-hmC in the ILPFC in response to learning. Extinction training led to a significant increase in TET3 messenger RNA levels within cortical neurons. TET3 was selectively activated within the adult neo-cortex in an experience-dependent manner.
A
Learning and memory have levels of permanence, differing from other mental processes such as thought, language, and consciousness, which are temporary in nature. Learning and memory can be either accumulated slowly (multiplication tables) or rapidly (touching a hot stove), but once attained, can be recalled into conscious use for a long time. Rats subjected to one instance of contextual fear conditioning create an especially strong long-term memory. At 24 hours after training, 9.17% of the genes in the genomes of rat hippocampus neurons were found to be differentially methylated. This included more than 2,000 differentially methylated genes at 24 hours after training, with over 500 genes being demethylated. Similar results to that in the rat hippocampus were also obtained in mice with contextual fear conditioning.
The hippocampus region of the brain is where contextual fear memories are first stored (see Figure), but this storage is transient and does not remain in the hippocampus. In rats contextual fear conditioning is abolished when the hippocampus is subjected to hippocampectomy just one day after conditioning, but rats retain a considerable amount of contextual fear when hippocampectomy is delayed by four weeks. In mice, examined at 4 weeks after conditioning, the hippocampus methylations and demethylations were reversed (the hippocampus is needed to form memories but memories are not stored there) while substantial differential CpG methylation and demethylation occurred in cortical neurons during memory maintenance. There were 1,223 differentially methylated genes in the anterior cingulate cortex (see Figure) of mice four weeks after contextual fear conditioning. Thus, while there were many methylations in the hippocampus shortly after memory was formed, all these hippocampus methylations were demethylated as soon as four weeks later.
Li et al. reported one example of the relationship between expression of a TET protein, demethylation and memory while using extinction training. Extinction training is the disappearance of a previously learned behavior when the behavior is not reinforced.
A comparison between infralimbic prefrontal cortex (ILPFC) neuron samples derived from mice trained to fear an auditory cue and extinction-trained mice revealed dramatic experience-dependent genome-wide differences in the accumulation of 5-hmC in the ILPFC in response to learning. Extinction training led to a significant increase in TET3 messenger RNA levels within cortical neurons. TET3 was selectively activated within the adult neo-cortex in an experience-dependent manner.
A
 The
The
DNA demethylation
For molecular biology in mammals, DNA demethylation causes replacement of 5-methylcytosine (5mC) in a DNA sequence by cytosine (C) (see figure of 5mC and C). DNA demethylation can occur by an active process at the site of a 5mC in a DNA sequenc ...
. 5-Methylcytosine (see first Figure) is a methylated
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These ...
form of the DNA base cytosine
Cytosine () ( symbol C or Cyt) is one of the four nucleobases found in DNA and RNA, along with adenine, guanine, and thymine (uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached (an am ...
(C) that often regulates gene transcription
Transcription refers to the process of converting sounds (voice, music etc.) into letters or musical notes, or producing a copy of something in another medium, including:
Genetics
* Transcription (biology), the copying of DNA into RNA, the fir ...
and has several other functions in the genome.
hydroxylation
In chemistry, hydroxylation can refer to:
*(i) most commonly, hydroxylation describes a chemical process that introduces a hydroxyl group () into an organic compound.
*(ii) the ''degree of hydroxylation'' refers to the number of OH groups in a ...
of DNA 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), and can further catalyse oxidation of 5hmC to 5-formylcytosine (5fC) and then to 5-carboxycytosine (5caC). 5fC and 5caC can be removed from the DNA base sequence by base excision repair
Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from t ...
and replaced by cytosine
Cytosine () ( symbol C or Cyt) is one of the four nucleobases found in DNA and RNA, along with adenine, guanine, and thymine (uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached (an am ...
in the base sequence.
DNA demethylation
For molecular biology in mammals, DNA demethylation causes replacement of 5-methylcytosine (5mC) in a DNA sequence by cytosine (C) (see figure of 5mC and C). DNA demethylation can occur by an active process at the site of a 5mC in a DNA sequenc ...
required during embryogenesis, gametogenesis, memory, learning, addiction and pain perception.
TET proteins
The three related ''TET'' genes, '' TET1'', ''TET2'' and ''TET3'' code respectively for three related mammalian proteins TET1, TET2, and TET3. All three proteins possess 5mC oxidase activity, but they differ in terms of domain architecture. TET proteins are large (∼180- to 230-kDa) multidomain enzymes. All TET proteins contain a conserved double-stranded β-helix (DSBH) domain, a cysteine-rich domain, and binding sites for the cofactors Fe(II) and 2-oxoglutarate (2-OG) that together form the core catalytic region in the C terminus. In addition to their catalytic domain, full-length TET1 and TET3 proteins have an N-terminal CXXC zinc finger domain that can bind DNA. The TET2 protein lacks a CXXC domain, but the ''IDAX'' gene, that's a neighbor of the TET2 gene, encodes a CXXC4 protein. IDAX is thought to play a role in regulating TET2 activity by facilitating its recruitment to unmethylated CpGs.TET isoforms
The three ''TET'' genes are expressed as differentisoforms
A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isof ...
, including at least two isoforms of TET1, three of TET2 and three of TET3. Different isoforms of the ''TET'' genes are expressed in different cells and tissues. The full-length canonical TET1 isoform appears virtually restricted to early embryos, embryonic stem cells and primordial germ cells (PGCs). The dominant TET1 isoform in most somatic tissues, at least in the mouse, arises from alternative promoter usage which gives rise to a short transcript and a truncated protein designated TET1s. The three isoforms of TET2 arise from different promoters. They are expressed and active in embryogenesis and differentiation of hematopoietic cells. The isoforms of TET3 are the full length form TET3FL, a short form splice variant TET3s, and a form that occurs in oocytes designated TET3o. TET3o is created by alternative promoter use and contains an additional first N-terminal exon coding for 11 amino acids. TET3o only occurs in oocytes and the one cell stage of the zygote and is not expressed in embryonic stem cells or in any other cell type or adult mouse tissue tested. Whereas TET1 expression can barely be detected in oocytes and zygotes, and TET2 is only moderately expressed, the TET3 variant TET3o shows extremely high levels of expression in oocytes and zygotes, but is nearly absent at the 2-cell stage. It appears that TET3o, high in oocytes and zygotes at the one cell stage, is the major TET enzyme utilized when almost 100% rapid demethylation occurs in the paternal genome just after fertilization and before DNA replication begins (see DNA demethylation
For molecular biology in mammals, DNA demethylation causes replacement of 5-methylcytosine (5mC) in a DNA sequence by cytosine (C) (see figure of 5mC and C). DNA demethylation can occur by an active process at the site of a 5mC in a DNA sequenc ...
).
TET specificity
Many different proteins bind to particular TET enzymes and recruit the TETs to specific genomic locations. In some studies, further analysis is needed to determine whether the interaction per se mediates the recruitment or instead the interacting partner helps to establish a favourable chromatin environment for TET binding. TET1‑depleted and TET2‑depleted cells revealed distinct target preferences of these two enzymes, with TET1‑preferring promoters and TET2‑preferring gene bodies of highly expressed genes and enhancers.DNA methyltransferase
In biochemistry, the DNA methyltransferase (DNA MTase, DNMT) family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl m ...
s (DNMTs) show a strong preference for adding a methyl group to the 5 carbon of a cytosine
Cytosine () ( symbol C or Cyt) is one of the four nucleobases found in DNA and RNA, along with adenine, guanine, and thymine (uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached (an am ...
where a cytosine nucleotide
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules wi ...
is followed by a guanine
Guanine () ( symbol G or Gua) is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine (uracil in RNA). In DNA, guanine is paired with cytosine. The guanine nucleoside is called ...
nucleotide in the linear sequence
In mathematics, a sequence is an enumerated collection of objects in which repetitions are allowed and order matters. Like a set, it contains members (also called ''elements'', or ''terms''). The number of elements (possibly infinite) is calle ...
of bases along its 5' → 3' direction (at CpG site
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG isl ...
s). This forms a 5mCpG site. More than 98% of DNA methylation occurs at CpG sites in mammalian somatic cell
A somatic cell (from Ancient Greek σῶμα ''sôma'', meaning "body"), or vegetal cell, is any biological cell forming the body of a multicellular organism other than a gamete, germ cell, gametocyte or undifferentiated stem cell. Such cells compo ...
s. Thus TET enzymes largely initiate demethylation at 5mCpG sites.
Oxoguanine glycosylase (OGG1) is one example of a protein that recruits a TET enzyme. TET1 is able to act on 5mCpG if an ROS has first acted on the guanine to form 8-hydroxy-2'-deoxyguanosine
8-Oxo-2'-deoxyguanosine (8-oxo-dG) is an oxidized derivative of deoxyguanosine. 8-Oxo-dG is one of the major products of DNA oxidation. Concentrations of 8-oxo-dG within a cell are a measurement of oxidative stress.
In DNA
Steady-state levels ...
(8-OHdG or its tautomer 8-oxo-dG), resulting in a 5mCp-8-OHdG dinucleotide (see Figure). After formation of 5mCp-8-OHdG, the base excision repair
Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from t ...
enzyme OGG1 binds to the 8-OHdG lesion without immediate excision (see Figure). Adherence of OGG1 to the 5mCp-8-OHdG site recruits TET1, allowing TET1 to oxidize the 5mC adjacent to 8-OHdG. This initiates the demethylation pathway.
EGR1
EGR-1 (Early growth response protein 1) also known as ZNF268 (zinc finger protein 268) or NGFI-A (nerve growth factor-induced protein A) is a protein that in humans is encoded by the ''EGR1'' gene.
EGR-1 is a mammalian transcription factor. It wa ...
is another example of a protein that recruits a TET enzyme. EGR1 has an important role in learning and memory. When a new event such as fear conditioning
Pavlovian fear conditioning is a behavioral paradigm in which organisms learn to predict aversive events. It is a form of learning in which an aversive stimulus (e.g. an electrical shock) is associated with a particular neutral context (e.g., a r ...
causes a memory to be formed, ''EGR1'' messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the p ...
is rapidly and selectively up-regulated in subsets of neurons in specific brain regions associated with learning and memory formation. TET1s is the predominant isoform of TET1 that is expressed in neurons. When EGR1 proteins are expressed, they appear to bring TET1s to about 600 sites in the neuron genome. Then EGR1 and TET1 appear to cooperate in demethylating and thereby activating the expression of genes downstream of the EGR1 binding sites in DNA.
TET processivity
TET processivity can be viewed at three levels, the physical, chemical and genetic levels. Physical processivity refers to the ability of a TET protein to slide along the DNA from one CpG site to another. An in vitro study showed that DNA-bound TET does not preferentially oxidize other CpG sites on the same DNA molecule, indicating that TET is not physically processive. Chemical processivity refers to the ability of TET to catalyze the oxidation of 5mC iteratively to 5caC without releasing its substrate. It appears that TET can work through both chemically processive and non‑processive mechanisms depending on reaction conditions. Genetic processivity refers to the genetic outcome of TET‑mediated oxidation in the genome, as shown by mapping of the oxidized bases. In mouse embryonic stem cells, many genomic regions orCpG site
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG isl ...
s are modified so that 5mC is changed to 5hmC but not to 5fC or 5caC, whereas at many otherCpG sites 5mCs are modified to 5fC or 5caC but not 5hmC, suggesting that 5mC is processed to different states at different genomic regions or CpG sites.
TET enzyme activity
TET enzymes aredioxygenase
Dioxygenases are oxidoreductase enzymes. Aerobic life, from simple single-celled bacteria species to complex eukaryotic organisms, has evolved to depend on the oxidizing power of dioxygen in various metabolic pathways. From energetic adenosine tri ...
s in the family of alpha-ketoglutarate-dependent hydroxylases Alpha-ketoglutarate-dependent hydroxylases are a major class of non-heme iron proteins that catalyse a wide range of reactions. These reactions include hydroxylation reactions, demethylations, ring expansions, ring closures, and desaturations. Func ...
. A TET enzyme is an alpha-ketoglutarate (α-KG) dependent dioxygenase that catalyses an oxidation reaction by incorporating a single oxygen atom from molecular oxygen (O2) into its substrate, 5-methylcytosine in DNA (5mC), to produce the product 5-hydroxymethylcytosine in DNA. This conversion is coupled with the oxidation of the co-substrate α-KG to succinate and carbon dioxide (see Figure).
The first step involves the binding of α-KG and 5-methylcytosine to the TET enzyme active site. The TET enzymes each harbor a core catalytic domain with a double-stranded β-helix fold that contains the crucial metal-binding residues found in the family of Fe(II)/α-KG- dependent oxygenases. α-KG coordinates as a bidentate ligand
In coordination chemistry, a ligand is an ion or molecule (functional group) that binds to a central metal atom to form a coordination complex. The bonding with the metal generally involves formal donation of one or more of the ligand's electr ...
(connected at two points) to Fe(II) (see Figure), while the 5mC is held by a noncovalent force in close proximity. The TET active site contains a highly conserved triad motif, in which the catalytically-essential Fe(II) is held by two histidine residues and one aspartic acid residue (see Figure). The triad binds to one face of the Fe center, leaving three labile sites available for binding α-KG and O2 (see Figure). TET then acts to convert 5-methylcytosine to 5-hydroxymethylcytosine while α-ketoglutarate is converted to succinate and CO2.
Alternate TET activities
The TET proteins also have activities that are independent of DNA demethylation. These include, for instance, TET2 interaction with O-linked N-acetylglucosamine ( O-GlcNAc) transferase to promote histone O-GlcN acylation to affect transcription of target genes.TET functions
Early embryogenesis
 The mouse
The mouse sperm
Sperm is the male reproductive cell, or gamete, in anisogamous forms of sexual reproduction (forms in which there is a larger, female reproductive cell and a smaller, male one). Animals produce motile sperm with a tail known as a flagellum, whi ...
genome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding ge ...
is 80–90% methylated
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These ...
at its CpG site
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG isl ...
s in DNA, amounting to about 20 million methylated sites. After fertilization
Fertilisation or fertilization (see spelling differences), also known as generative fertilisation, syngamy and impregnation, is the fusion of gametes to give rise to a new individual organism or offspring and initiate its development. Proce ...
, early in the first day of embryogenesis
An embryo is an initial stage of development of a multicellular organism. In organisms that reproduce sexually, embryonic development is the part of the life cycle that begins just after fertilization of the female egg cell by the male sperm ...
, the paternal chromosomes are almost completely demethylated Demethylation is the chemical process resulting in the removal of a methyl group (CH3) from a molecule. A common way of demethylation is the replacement of a methyl group by a hydrogen atom, resulting in a net loss of one carbon and two hydrogen ato ...
in six hours by an active TET-dependent process, before DNA replication begins (blue line in Figure).
Demethylation of the maternal genome occurs by a different process. In the mature oocyte
An oocyte (, ), oöcyte, or ovocyte is a female gametocyte or germ cell involved in reproduction. In other words, it is an immature ovum, or egg cell. An oocyte is produced in a female fetus in the ovary during female gametogenesis. The female ...
, about 40% of its CpG sites in DNA are methylated. In the pre-implantation embryo up to the blastocyst stage (see Figure), the only methyltransferase present is an isoform of DNMT1
DNA (cytosine-5)-methyltransferase 1 is an enzyme that catalyzes the transfer of methyl groups to specific CpG structures in DNA, a process called DNA methylation. In humans, it is encoded by the ''DNMT1'' gene. DNMT1 forms part of the family of ...
designated DNMT1o. It appears that demethylation of the maternal chromosomes largely takes place by blockage of the methylating enzyme DNMT1o from entering the nucleus except briefly at the 8 cell stage (see DNA demethylation
For molecular biology in mammals, DNA demethylation causes replacement of 5-methylcytosine (5mC) in a DNA sequence by cytosine (C) (see figure of 5mC and C). DNA demethylation can occur by an active process at the site of a 5mC in a DNA sequenc ...
). The maternal-origin DNA thus undergoes passive demethylation by dilution of the methylated maternal DNA during replication (red line in Figure). The morula
A morula (Latin, ''morus'': mulberry) is an early-stage embryo consisting of a solid ball of cells called blastomeres, contained in mammals, and other animals within the zona pellucida shell. The blastomeres are the daughter cells of the zygote ...
(at the 16 cell stage), has only a small amount of DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts t ...
(black line in Figure).
Gametogenesis
The newly formed primordial germ cells (PGC) in the implanted embryo devolve from the somatic cells at about day 7 of embryogenesis in the mouse. At this point the PGCs have high levels of methylation. These cells migrate from the epiblast toward thegonadal ridge
The genital ridge (or gonadal ridge) is the precursor to the gonads. The genital ridge initially consists mainly of mesenchyme and cells of underlying mesonephric origin. Once oogonia enter this area they attempt to associate with these somatic cel ...
. As reviewed by Messerschmidt et al., the majority of PGCs are arrested in the G2 phase of the cell cycle while they migrate toward the hindgut during embryo days 7.5 to 8.5. Then demethylation of the PGCs takes place in two waves. There is both passive and active, TET-dependent demethylation of the primordial germ cells. At day 9.5 the primordial germ cells begin to rapidly replicate going from about 200 PGCs at embryo day 9.5 to about 10,000 PGCs at day 12.5. During days 9.5 to 12.5 DNMT3a and DNMT3b are repressed and DNMT1 is present in the nucleus at a high level. But DNMT1 is unable to methylate cytosines during days 9.5 to 12.5 because the ''UHRF1
Ubiquitin-like, containing PHD and RING finger domains, 1, also known as UHRF1, is a protein which in humans is encoded by the ''UHRF1'' gene.
Function
This gene encodes a member of a subfamily of RING finger domain, RING-finger type E3 ubiqui ...
'' gene (also known as ''NP95'') is repressed and UHRF1 is an essential protein needed to recruit DNMT1 to replication foci where maintenance DNA methylation takes place. This is a passive, dilution form of demethylation.
In addition, from embryo day 9.5 to 13.5 there is an active form of demethylation. As indicated in the Figure of the demethylation pathway above, two enzymes are central to active demethylation. These are a ten-eleven translocation (TET) methylcytosine dioxygenase and thymine-DNA glycosylase
G/T mismatch-specific thymine DNA glycosylase is an enzyme that in humans is encoded by the TDG gene. Several bacterial proteins have strong sequence homology with this protein.
Function
The protein encoded by this gene belongs to the TDG/mug D ...
(TDG). One particular TET enzyme, TET1, and TDG are present at high levels from embryo day 9.5 to 13.5, and are employed in active TET-dependent demethylation during gametogenesis. PGC genomes display the lowest levels of DNA methylation of any cells in the entire life cycle of the mouse by embryonic day 13.5.
Learning and Memory
 Learning and memory have levels of permanence, differing from other mental processes such as thought, language, and consciousness, which are temporary in nature. Learning and memory can be either accumulated slowly (multiplication tables) or rapidly (touching a hot stove), but once attained, can be recalled into conscious use for a long time. Rats subjected to one instance of contextual fear conditioning create an especially strong long-term memory. At 24 hours after training, 9.17% of the genes in the genomes of rat hippocampus neurons were found to be differentially methylated. This included more than 2,000 differentially methylated genes at 24 hours after training, with over 500 genes being demethylated. Similar results to that in the rat hippocampus were also obtained in mice with contextual fear conditioning.
The hippocampus region of the brain is where contextual fear memories are first stored (see Figure), but this storage is transient and does not remain in the hippocampus. In rats contextual fear conditioning is abolished when the hippocampus is subjected to hippocampectomy just one day after conditioning, but rats retain a considerable amount of contextual fear when hippocampectomy is delayed by four weeks. In mice, examined at 4 weeks after conditioning, the hippocampus methylations and demethylations were reversed (the hippocampus is needed to form memories but memories are not stored there) while substantial differential CpG methylation and demethylation occurred in cortical neurons during memory maintenance. There were 1,223 differentially methylated genes in the anterior cingulate cortex (see Figure) of mice four weeks after contextual fear conditioning. Thus, while there were many methylations in the hippocampus shortly after memory was formed, all these hippocampus methylations were demethylated as soon as four weeks later.
Li et al. reported one example of the relationship between expression of a TET protein, demethylation and memory while using extinction training. Extinction training is the disappearance of a previously learned behavior when the behavior is not reinforced.
A comparison between infralimbic prefrontal cortex (ILPFC) neuron samples derived from mice trained to fear an auditory cue and extinction-trained mice revealed dramatic experience-dependent genome-wide differences in the accumulation of 5-hmC in the ILPFC in response to learning. Extinction training led to a significant increase in TET3 messenger RNA levels within cortical neurons. TET3 was selectively activated within the adult neo-cortex in an experience-dependent manner.
A
Learning and memory have levels of permanence, differing from other mental processes such as thought, language, and consciousness, which are temporary in nature. Learning and memory can be either accumulated slowly (multiplication tables) or rapidly (touching a hot stove), but once attained, can be recalled into conscious use for a long time. Rats subjected to one instance of contextual fear conditioning create an especially strong long-term memory. At 24 hours after training, 9.17% of the genes in the genomes of rat hippocampus neurons were found to be differentially methylated. This included more than 2,000 differentially methylated genes at 24 hours after training, with over 500 genes being demethylated. Similar results to that in the rat hippocampus were also obtained in mice with contextual fear conditioning.
The hippocampus region of the brain is where contextual fear memories are first stored (see Figure), but this storage is transient and does not remain in the hippocampus. In rats contextual fear conditioning is abolished when the hippocampus is subjected to hippocampectomy just one day after conditioning, but rats retain a considerable amount of contextual fear when hippocampectomy is delayed by four weeks. In mice, examined at 4 weeks after conditioning, the hippocampus methylations and demethylations were reversed (the hippocampus is needed to form memories but memories are not stored there) while substantial differential CpG methylation and demethylation occurred in cortical neurons during memory maintenance. There were 1,223 differentially methylated genes in the anterior cingulate cortex (see Figure) of mice four weeks after contextual fear conditioning. Thus, while there were many methylations in the hippocampus shortly after memory was formed, all these hippocampus methylations were demethylated as soon as four weeks later.
Li et al. reported one example of the relationship between expression of a TET protein, demethylation and memory while using extinction training. Extinction training is the disappearance of a previously learned behavior when the behavior is not reinforced.
A comparison between infralimbic prefrontal cortex (ILPFC) neuron samples derived from mice trained to fear an auditory cue and extinction-trained mice revealed dramatic experience-dependent genome-wide differences in the accumulation of 5-hmC in the ILPFC in response to learning. Extinction training led to a significant increase in TET3 messenger RNA levels within cortical neurons. TET3 was selectively activated within the adult neo-cortex in an experience-dependent manner.
A short hairpin RNA
A short hairpin RNA or small hairpin RNA (shRNA/Hairpin Vector) is an artificial RNA molecule with a tight hairpin turn that can be used to silence target gene expression via RNA interference (RNAi). Expression of shRNA in cells is typically acc ...
(shRNA) is an artificial RNA
Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid ( DNA) are nucleic acids. Along with lipids, proteins, and carbohydra ...
molecule with a tight hairpin turn that can be used to silence target gene expression via RNA interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by o ...
. Mice trained in the presence of TET3-targeted shRNA showed a significant impairment in fear extinction memory.
Addiction
 The
The nucleus accumbens
The nucleus accumbens (NAc or NAcc; also known as the accumbens nucleus, or formerly as the ''nucleus accumbens septi'', Latin for "nucleus adjacent to the septum") is a region in the basal forebrain rostral to the preoptic area of the hypotha ...
(NAc) has a significant role in addiction
Addiction is a neuropsychological disorder characterized by a persistent and intense urge to engage in certain behaviors, one of which is the usage of a drug, despite substantial harm and other negative consequences. Repetitive drug use o ...
. In the nucleus accumbens of mice, repeated cocaine exposure resulted in reduced ''TET1'' messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the p ...
(mRNA) and reduced TET1 protein expression. Similarly, there was a ~40% decrease in ''TET1'' mRNA in the NAc of human cocaine addicts examined postmortem.
As indicated above in learning and memory, a short hairpin RNA
A short hairpin RNA or small hairpin RNA (shRNA/Hairpin Vector) is an artificial RNA molecule with a tight hairpin turn that can be used to silence target gene expression via RNA interference (RNAi). Expression of shRNA in cells is typically acc ...
(shRNA) is an artificial RNA
Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid ( DNA) are nucleic acids. Along with lipids, proteins, and carbohydra ...
molecule with a tight hairpin turn that can be used to silence target gene expression via RNA interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by o ...
. Feng et al. injected shRNA targeted to ''TET1'' in the NAc of mice. This could reduce ''TET1'' expression in the same manner as reduction of ''TET1'' expression with cocaine exposure. They then used an indirect measure of addiction, conditioned place preference
Conditioned place preference (CPP) is a form of Pavlovian conditioning used to measure the motivational effects of objects or experiences. This motivation comes from the pleasurable aspect of the experience, so that the brain can be reminded of th ...
. Conditioned place preference can measure the amount of time an animal spends in an area that has been associated with cocaine exposure, and this can indicate an addiction to cocaine. Reduced ''Tet1'' expression caused by shRNA injected into the NAc robustly enhanced cocaine place conditioning.
Pain (Nociception)
As described in the articleNociception
Nociception (also nocioception, from Latin ''nocere'' 'to harm or hurt') is the sensory nervous system's process of encoding noxious stimuli. It deals with a series of events and processes required for an organism to receive a painful stimulus, co ...
, nociception is the sensory nervous system's response to harmful stimuli, such as a toxic chemical applied to a tissue. In nociception, chemical stimulation of sensory nerve cells called nociceptor
A nociceptor ("pain receptor" from Latin ''nocere'' 'to harm or hurt') is a sensory neuron that responds to damaging or potentially damaging stimuli by sending "possible threat" signals to the spinal cord and the brain. The brain creates the sens ...
s produces a signal that travels along a chain of nerve fibers via the spinal cord
The spinal cord is a long, thin, tubular structure made up of nervous tissue, which extends from the medulla oblongata in the brainstem to the lumbar region of the vertebral column (backbone). The backbone encloses the central canal of the spi ...
to the brain
A brain is an organ that serves as the center of the nervous system in all vertebrate and most invertebrate animals. It is located in the head, usually close to the sensory organs for senses such as vision. It is the most complex organ in a v ...
. Nociception triggers a variety of physiological and behavioral responses and usually results in a subjective experience, or perception
Perception () is the organization, identification, and interpretation of sensory information in order to represent and understand the presented information or environment. All perception involves signals that go through the nervous system ...
, of pain
Pain is a distressing feeling often caused by intense or damaging stimuli. The International Association for the Study of Pain defines pain as "an unpleasant sensory and emotional experience associated with, or resembling that associated with, ...
.
Work by Pan et al.{{cite journal , vauthors=Pan Z, Zhang M, Ma T, Xue ZY, Li GF, Hao LY, Zhu LJ, Li YQ, Ding HL, Cao JL , title=Hydroxymethylation of microRNA-365-3p Regulates Nociceptive Behaviors via Kcnh2 , journal=J. Neurosci. , volume=36 , issue=9 , pages=2769–81 , date=March 2016 , pmid=26937014 , pmc=6604871 , doi=10.1523/JNEUROSCI.3474-15.2016 first showed that TET1 and TET3 proteins are normally present in the spinal cords of mice. They used a pain inducing model of intra plantar
Standard anatomical terms of location are used to unambiguously describe the anatomy of animals, including humans. The terms, typically derived from Latin or Greek roots, describe something in its standard anatomical position. This position prov ...
injection of 5% formalin into the dorsal surface of the mouse hindpaw and measured time of licking of the hindpaw as a measure of induced pain. Protein expression of TET1 and TET3 increased by 152% and 160%, respectively, by 2 hours after formalin injection. Forced reduction of expression of TET1 or TET3 by spinal injection of Tet1-siRNA or Tet3-siRNA for three consecutive days before formalin injection alleviated the mouse perception of pain. On the other hand, forced overexpression of TET1 or TET3 for 2 consecutive days significantly produced pain-like behavior as evidenced by a decrease in the mouse of the thermal pain threshold.
They further showed that the nociceptive pain effects occurred through TET mediated conversion of 5-methylcytosine to 5-hydroxymethylcytosine in the promoter of a microRNA
MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides. Found in plants, animals and some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRN ...
designated miR-365-3p, thus increasing its expression. This microRNA, in turn, ordinarily targets (decreases expression of) the messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the p ...
of ''Kcnh2'', that codes for a protein known as Kv11.1 or KCNH2. KCNH2 is the alpha subunit of a potassium ion channel
Potassium channels are the most widely distributed type of ion channel found in virtually all organisms. They form potassium-selective pores that span cell membranes. Potassium channels are found in most cell types and control a wide variety of ce ...
in the central nervous system. Forced decrease in expression of TET1 or TET3 through pre-injection of siRNA reversed the decrease of KCNH2 protein in formalin-treated mice.
References