inverted repeat on:
[Wikipedia]
[Google]
[Amazon]
An inverted repeat (or IR) is a single stranded sequence of
 Beginning with this initial sequence:
Beginning with this initial sequence:
The complement created by base pairing is:
The reverse complement is:
And, the inverted repeat sequence is:
"nnnnnn" represents any number of intervening nucleotides.
EXAMPLE:
Step 1: start with an inverted repeat:
Step 2: remove intervening nucleotides:
This resulting sequence is palindromic because it is the reverse complement of itself. ::: test sequence (from Step 2 with intervening nucleotides removed) ::: complement of test sequence ::: reverse complement This is the same as the test sequence above, and thus, it is a palindrome.
 For quantification and comparison of inverted repeats between several species, namely on archaea, see
For quantification and comparison of inverted repeats between several species, namely on archaea, see
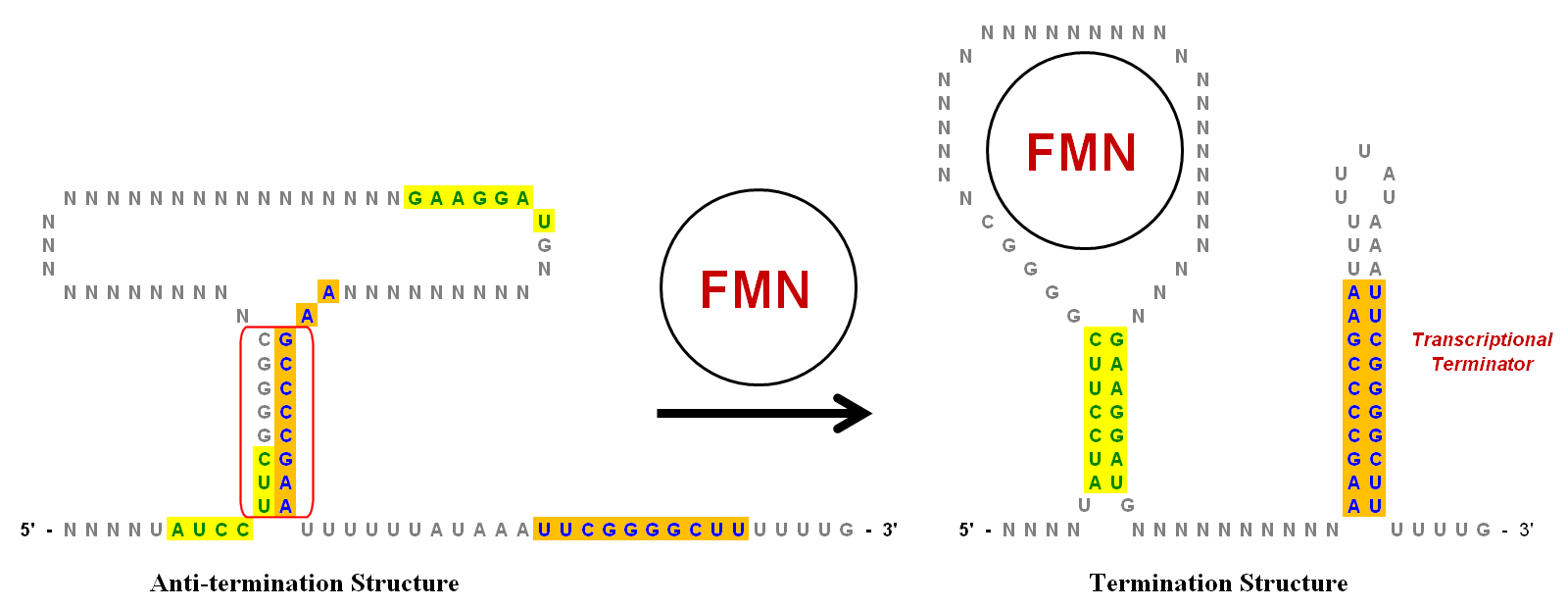 In the absence of FMN, the Anti-termination structure is the preferred conformation for the mRNA transcript. It is created by base-pairing of the inverted repeat region circled in red. When FMN is present, it may bind to the loop and prevent formation of the Anti-termination structure. This allows two different sets of inverted repeats to base-pair and form the Termination structure. The stem-loop on the 3' end is a transcriptional terminator because the sequence immediately following it is a string of uracils (U). If this stem-loop forms (due to the presence of FMN) as the growing RNA strand emerges from the
In the absence of FMN, the Anti-termination structure is the preferred conformation for the mRNA transcript. It is created by base-pairing of the inverted repeat region circled in red. When FMN is present, it may bind to the loop and prevent formation of the Anti-termination structure. This allows two different sets of inverted repeats to base-pair and form the Termination structure. The stem-loop on the 3' end is a transcriptional terminator because the sequence immediately following it is a string of uracils (U). If this stem-loop forms (due to the presence of FMN) as the growing RNA strand emerges from the
 The illustration shows an inverted repeat undergoing cruciform extrusion. DNA in the region of the inverted repeat unwinds and then recombines, forming a four-way junction with two
The illustration shows an inverted repeat undergoing cruciform extrusion. DNA in the region of the inverted repeat unwinds and then recombines, forming a four-way junction with two 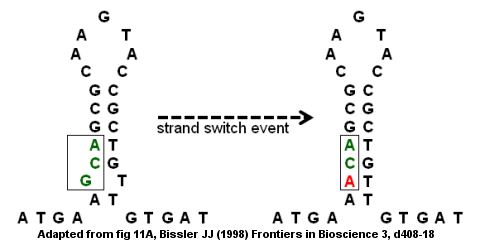

non-B DB
A Database for Integrated Annotations and Analysis of non-B DNA Forming Motifs. This database is provided by The Advanced Biomedical Computing Center (ABCC) at then Frederick National Laboratory for Cancer Research (FNLCR). It covers the A-DNA and Z-DNA conformations otherwise known as "non-B DNAs" because they are not the more common B-DNA form of a right-handed Watson-Crick
Inverted Repeats Database
Boston University. This database is a web application that allows query and analysis of repeats held in the PUBLIC DATABASE project. Scientists can also analyze their own sequences with the Inverted Repeats Finder algorithm.
P-MITE: a Plant MITE database
— this database for
EMBOSS website
Applications specifically related to inverted repeats are listed below:
Finds inverted repeats in nucleotide sequences. Threshold values can be set to limit the scope of the search.
EMBOSS palindrome
Finds
nucleotide
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both o ...
s followed downstream by its reverse complement. The intervening sequence of nucleotides between the initial sequence and the reverse complement can be any length including zero. For example, is an inverted repeat sequence. When the intervening length is zero, the composite sequence is a palindromic sequence
A palindromic sequence is a nucleic acid sequence in a double-stranded DNA or RNA molecule whereby reading in a certain direction (e.g. 5' to 3') on one strand is identical to the sequence in the same direction (e.g. 5' to 3') on the complemen ...
.
Both inverted repeats and direct repeats constitute types of nucleotide sequences that occur repetitively. These repeated DNA sequences often range from a pair of nucleotides to a whole gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
, while the proximity of the repeat sequences varies between widely dispersed and simple tandem arrays. The short tandem repeat
In genetics, tandem repeats occur in DNA when a pattern of one or more nucleotides is repeated and the repetitions are directly adjacent to each other, e.g. ATTCG ATTCG ATTCG, in which the sequence ATTCG is repeated three times.
Several protein ...
sequences may exist as just a few copies in a small region to thousands of copies dispersed all over the genome of most eukaryotes
The eukaryotes ( ) constitute the domain of Eukaryota or Eukarya, organisms whose cells have a membrane-bound nucleus. All animals, plants, fungi, seaweeds, and many unicellular organisms are eukaryotes. They constitute a major group of ...
. Repeat sequences with about 10–100 base pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
s are known as minisatellite
In genetics, a minisatellite is a tract of repetitive DNA in which certain DNA motifs (ranging in length from 10–60 base pairs) are typically repeated two to several hundred times. Minisatellites occur at more than 1,000 locations in the huma ...
s, while shorter repeat sequences having mostly 2–4 base pairs are known as microsatellite
A microsatellite is a tract of repetitive DNA in which certain Sequence motif, DNA motifs (ranging in length from one to six or more base pairs) are repeated, typically 5–50 times. Microsatellites occur at thousands of locations within an organ ...
s. The most common repeats include the dinucleotide repeats, which have the bases AC on one DNA strand, and GT on the complementary strand. Some elements of the genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
with unique sequences function as exons
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence i ...
, introns
An intron is any Nucleic acid sequence, nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of ...
and regulatory DNA. Though the most familiar loci of the repetitive sequences are the centromere
The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fiber ...
and the telomere
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes (see #Sequences, Sequences). Telomeres are a widespread genetic feature most commonly found in eukaryotes. In ...
, a large portion of the repeated sequences in the genome are found among the noncoding DNA
Non-coding DNA (ncDNA) sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA is transcribed into functional non-coding RNA molecules (e.g. transfer RNA, microRNA, piRNA, ribosomal RNA, and regu ...
.
Inverted repeats have a number of important biological functions. They define the boundaries in transposon
A transposable element (TE), also transposon, or jumping gene, is a type of mobile genetic element, a nucleic acid sequence in DNA that can change its position within a genome.
The discovery of mobile genetic elements earned Barbara McClinto ...
s and indicate regions capable of self-complementary base pairing
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
(regions within a single sequence which can base pair with each other). These properties play an important role in genome instability and contribute not only to cellular evolution and genetic diversity
Genetic diversity is the total number of genetic characteristics in the genetic makeup of a species. It ranges widely, from the number of species to differences within species, and can be correlated to the span of survival for a species. It is d ...
but also to mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, ...
and disease
A disease is a particular abnormal condition that adversely affects the structure or function (biology), function of all or part of an organism and is not immediately due to any external injury. Diseases are often known to be medical condi ...
. In order to study these effects in detail, a number of programs and databases have been developed to assist in discovery and annotation of inverted repeats in various genomes.
Understanding inverted repeats
Example of an inverted repeat
 Beginning with this initial sequence:
Beginning with this initial sequence:The complement created by base pairing is:
The reverse complement is:
And, the inverted repeat sequence is:
"nnnnnn" represents any number of intervening nucleotides.
Vs. direct repeat
A direct repeat occurs when a sequence is repeated with the same pattern downstream. There is no inversion and no reverse complement associated with a direct repeat. The nucleotide sequence written in bold characters signifies the repeated sequence. It may or may not have intervening nucleotides. ::: ::: Linguistically, a typical direct repeat is comparable to rhyming, as in "time on a dime".Vs. tandem repeat
A direct repeat with ''no'' intervening nucleotides between the initial sequence and its downstream copy is aTandem repeat
In genetics, tandem repeats occur in DNA when a pattern of one or more nucleotides is repeated and the repetitions are directly adjacent to each other, e.g. ATTCG ATTCG ATTCG, in which the sequence ATTCG is repeated three times.
Several protein ...
. The nucleotide sequence written in bold characters signifies the repeated sequence.
:::
:::
Linguistically, a typical tandem repeat is comparable to stuttering, or deliberately repeated words, as in "bye-bye".
Vs. palindrome
An inverted repeat sequence with ''no'' intervening nucleotides between the initial sequence and its downstream reverse complement is apalindrome
A palindrome (Help:IPA/English, /ˈpæl.ɪn.droʊm/) is a word, palindromic number, number, phrase, or other sequence of symbols that reads the same backwards as forwards, such as ''madam'' or ''racecar'', the date "Twosday, 02/02/2020" and th ...
. EXAMPLE:
Step 1: start with an inverted repeat:
Step 2: remove intervening nucleotides:
This resulting sequence is palindromic because it is the reverse complement of itself. ::: test sequence (from Step 2 with intervening nucleotides removed) ::: complement of test sequence ::: reverse complement This is the same as the test sequence above, and thus, it is a palindrome.
Biological features and functionality
Conditions that favor synthesis
The diverse genome-wide repeats are derived fromtransposable element
A transposable element (TE), also transposon, or jumping gene, is a type of mobile genetic element, a nucleic acid sequence in DNA that can change its position within a genome.
The discovery of mobile genetic elements earned Barbara McClinto ...
s, which are now understood to "jump" about different genomic locations, without transferring their original copies. Subsequent shuttling of the same sequences over numerous generations ensures their multiplicity throughout the genome. The limited recombination of the sequences between two distinct sequence elements known as conservative site-specific recombination (CSSR) results in inversions of the DNA segment, based on the arrangement of the recombination recognition sequences on the donor DNA and recipient DNA. Again, the orientation of two of the recombining sites within the donor DNA molecule relative to the asymmetry of the intervening DNA cleavage sequences, known as the crossover region, is pivotal to the formation of either inverted repeats or direct repeats. Thus, recombination occurring at a pair of inverted sites will invert the DNA sequence between the two sites. Very stable chromosomes have been observed with comparatively fewer numbers of inverted repeats than direct repeats, suggesting a relationship between chromosome stability and the number of repeats.
Regions where presence is obligatory
Terminal inverted repeats have been observed in the DNA of various eukaryotic transposons, even though their source remains unknown. Inverted repeats are principally found at the origins of replication of cell organism and organelles that range from phage plasmids, mitochondria, and eukaryotic viruses to mammalian cells. The replication origins of the phage G4 and other related phages comprise a segment of nearly 139 nucleotide bases that include three inverted repeats that are essential for replication priming.In the genome
To a large extent, portions of nucleotide repeats are quite often observed as part of rare DNA combinations. The three main repeats which are largely found in particular DNA constructs include the closely precise homopurine-homopyrimidine inverted repeats, which is otherwise referred to as H palindromes, a common occurrence in triple helical H conformations that may comprise either the TAT or CGC nucleotide triads. The others could be described as long inverted repeats having the tendency to produce hairpins and cruciform, and finally direct tandem repeats, which commonly exist in structures described as slipped-loop, cruciform and left-handed Z-DNA.Common in different organisms
Past studies suggest that repeats are a common feature ofeukaryotes
The eukaryotes ( ) constitute the domain of Eukaryota or Eukarya, organisms whose cells have a membrane-bound nucleus. All animals, plants, fungi, seaweeds, and many unicellular organisms are eukaryotes. They constitute a major group of ...
unlike the prokaryotes
A prokaryote (; less commonly spelled procaryote) is a single-celled organism whose cell lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Ancient Greek (), meaning 'before', and (), meaning 'nut' ...
and archaea
Archaea ( ) is a Domain (biology), domain of organisms. Traditionally, Archaea only included its Prokaryote, prokaryotic members, but this has since been found to be paraphyletic, as eukaryotes are known to have evolved from archaea. Even thou ...
. Other reports suggest that irrespective of the comparative shortage of repeat elements in prokaryotic genomes, they nevertheless contain hundreds or even thousands of large repeats. Current genomic analysis seem to suggest the existence of a large excess of perfect inverted repeats in many prokaryotic genomes as compared to eukaryotic genomes.
 For quantification and comparison of inverted repeats between several species, namely on archaea, see
For quantification and comparison of inverted repeats between several species, namely on archaea, see
Inverted repeats in pseudoknots
Pseudoknot
__NOTOC__
A pseudoknot is a nucleic acid secondary structure containing at least two stem-loop structures in which half of one stem is intercalated between the two halves of another stem. The pseudoknot was first recognized in the turnip yellow ...
s are common structural motifs found in RNA. They are formed by two nested stem-loop
Stem-loops are nucleic acid Biomolecular structure, secondary structural elements which form via intramolecular base pairing in single-stranded DNA or RNA. They are also referred to as hairpins or hairpin loops. A stem-loop occurs when two regi ...
s such that the stem of one structure is formed from the loop of the other. There are multiple folding topologies among pseudoknots and great variation in loop lengths, making them a structurally diverse group.
Inverted repeats are a key component of pseudoknots as can be seen in the illustration of a naturally occurring pseudoknot found in the human telomerase RNA component. Four different sets of inverted repeats are involved in this structure. Sets 1 and 2 are the stem of stem-loop A and are part of the loop for stem-loop B. Similarly, sets 3 and 4 are the stem for stem-loop B and are part of the loop for stem-loop A.
Pseudoknots play a number of different roles in biology. The telomerase pseudoknot in the illustration is critical to that enzyme's activity. The ribozyme
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to Catalysis, catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozy ...
for the ''hepatitis delta virus (HDV)'' folds into a double-pseudoknot structure and self-cleaves its circular genome to produce a single-genome-length RNA. Pseudoknots also play a role in programmed ribosomal frameshifting found in some viruses and required in the replication of retroviruses
A retrovirus is a type of virus that inserts a DNA copy of its RNA genome into the DNA of a host cell that it invades, thus changing the genome of that cell. After invading a host cell's cytoplasm, the virus uses its own reverse transcriptase ...
.
In riboswitches
Inverted repeats play an important role inriboswitch
In molecular biology, a riboswitch is a regulatory segment of a messenger RNA molecule that binds a small molecule, resulting in a change in Translation (biology), production of the proteins encoded by the mRNA. Thus, an mRNA that contains a ribo ...
es, which are RNA regulatory elements that control the expression of genes that produce the mRNA, of which they are part. A simplified example of the flavin mononucleotide
Flavin mononucleotide (FMN), or riboflavin-5′-phosphate, is a biomolecule produced from riboflavin (vitamin B2) by the enzyme riboflavin kinase and functions as the prosthetic group of various oxidoreductases, including NADH dehydrogenase, as ...
(FMN) riboswitch is shown in the illustration. This riboswitch exists in the mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
transcript and has several stem-loop
Stem-loops are nucleic acid Biomolecular structure, secondary structural elements which form via intramolecular base pairing in single-stranded DNA or RNA. They are also referred to as hairpins or hairpin loops. A stem-loop occurs when two regi ...
structures upstream from the coding region
The coding region of a gene, also known as the coding DNA sequence (CDS), is the portion of a gene's DNA or RNA that codes for a protein. Studying the length, composition, regulation, splicing, structures, and functions of coding regions compared ...
. However, only the key stem-loops are shown in the illustration, which has been greatly simplified to help show the role of the inverted repeats. There are multiple inverted repeats in this riboswitch as indicated in green (yellow background) and blue (orange background).  In the absence of FMN, the Anti-termination structure is the preferred conformation for the mRNA transcript. It is created by base-pairing of the inverted repeat region circled in red. When FMN is present, it may bind to the loop and prevent formation of the Anti-termination structure. This allows two different sets of inverted repeats to base-pair and form the Termination structure. The stem-loop on the 3' end is a transcriptional terminator because the sequence immediately following it is a string of uracils (U). If this stem-loop forms (due to the presence of FMN) as the growing RNA strand emerges from the
In the absence of FMN, the Anti-termination structure is the preferred conformation for the mRNA transcript. It is created by base-pairing of the inverted repeat region circled in red. When FMN is present, it may bind to the loop and prevent formation of the Anti-termination structure. This allows two different sets of inverted repeats to base-pair and form the Termination structure. The stem-loop on the 3' end is a transcriptional terminator because the sequence immediately following it is a string of uracils (U). If this stem-loop forms (due to the presence of FMN) as the growing RNA strand emerges from the RNA polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.
Using the e ...
complex, it will create enough structural tension to cause the RNA strand to dissociate and thus terminate transcription. The dissociation occurs easily because the base-pairing between the U's in the RNA and the A's in the template strand are the weakest of all base-pairings. Thus, at higher concentration levels, FMN down-regulates its own transcription by increasing the formation of the termination structure.
Mutations and disease
Inverted repeats are often described as "hotspots" of eukaryotic and prokaryotic genomic instability. Long inverted repeats are deemed to greatly influence the stability of the genome of various organisms. This is exemplified in ''E. coli'', where genomic sequences with long inverted repeats are seldom replicated, but rather deleted with rapidity. Again, the long inverted repeats observed in yeast greatly favor recombination within the same and adjacent chromosomes, resulting in an equally very high rate of deletion. Finally, a very high rate of deletion and recombination were also observed in mammalian chromosomes regions with inverted repeats. Reported differences in the stability of genomes of interrelated organisms are always an indication of a disparity in inverted repeats. The instability results from the tendency of inverted repeats to fold into hairpin- or cruciform-like DNA structures. These special structures can hinder or confuse DNA replication and other genomic activities. Thus, inverted repeats lead to special configurations in bothRNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
and DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
that can ultimately cause mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, ...
s and disease
A disease is a particular abnormal condition that adversely affects the structure or function (biology), function of all or part of an organism and is not immediately due to any external injury. Diseases are often known to be medical condi ...
.
stem-loop
Stem-loops are nucleic acid Biomolecular structure, secondary structural elements which form via intramolecular base pairing in single-stranded DNA or RNA. They are also referred to as hairpins or hairpin loops. A stem-loop occurs when two regi ...
structures. The cruciform structure occurs because the inverted repeat sequences self-pair to each other on their own strand.
Extruded cruciforms can lead to frameshift mutation
A frameshift mutation (also called a framing error or a reading frame shift) is a genetic mutation caused by indels ( insertions or deletions) of a number of nucleotides in a DNA sequence that is not divisible by three. Due to the triplet natur ...
s when a DNA sequence has inverted repeats in the form of a palindrome
A palindrome (Help:IPA/English, /ˈpæl.ɪn.droʊm/) is a word, palindromic number, number, phrase, or other sequence of symbols that reads the same backwards as forwards, such as ''madam'' or ''racecar'', the date "Twosday, 02/02/2020" and th ...
combined with regions of direct repeats on either side. During transcription, slippage and partial dissociation of the polymerase from the template strand can lead to both deletion and insertion mutations. Deletion occurs when a portion of the unwound template strand
Transcription is the process of copying a segment of DNA into RNA for the purpose of gene expression. Some segments of DNA are transcribed into RNA molecules that can encode proteins, called messenger RNA (mRNA). Other segments of DNA are transc ...
forms a stem-loop that gets "skipped" by the transcription machinery. Insertion occurs when a stem-loop forms in a dissociated portion of the nascent (newly synthesized) strand causing a portion of the template strand to be transcribed twice.

Antithrombin deficiency from a point mutation
Imperfect inverted repeats can lead tomutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, ...
s through intrastrand and interstrand switching. The antithrombin
Antithrombin (AT) is a small glycoprotein that inactivates several enzymes of the coagulation system. It is a 464-amino-acid protein produced by the liver. It contains three disulfide bonds and a total of four possible glycosylation sites. α-An ...
III gene's coding region is an example of an imperfect inverted repeat as shown in the figure on the right.
The stem-loop
Stem-loops are nucleic acid Biomolecular structure, secondary structural elements which form via intramolecular base pairing in single-stranded DNA or RNA. They are also referred to as hairpins or hairpin loops. A stem-loop occurs when two regi ...
structure forms with a bump at the bottom because the G and T do not pair up. A strand switch event could result in the G (in the bump) being replaced by an A which removes the "imperfection" in the inverted repeat and provides a stronger stem-loop structure. However, the replacement also creates a point mutation
A point mutation is a genetic mutation where a single nucleotide base is changed, inserted or deleted from a DNA or RNA sequence of an organism's genome. Point mutations have a variety of effects on the downstream protein product—consequences ...
converting the GCA codon to ACA. If the strand switch event is followed by a second round of DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biolog ...
, the mutation may become fixed in the genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
and lead to disease. Specifically, the missense mutation
In genetics, a missense mutation is a point mutation in which a single nucleotide change results in a codon that codes for a different amino acid. It is a type of nonsynonymous substitution. Missense mutations change amino acids, which in turn alt ...
would lead to a defective gene and a deficiency in antithrombin which could result in the development of venous thromboembolism
Venous thrombosis is the blockage of a vein caused by a thrombus (blood clot). A common form of venous thrombosis is deep vein thrombosis (DVT), when a blood clot forms in the deep veins. If a thrombus breaks off ( embolizes) and flows to the lun ...
(blood clots within a vein).

Osteogenesis imperfecta from a frameshift mutation
Mutations in thecollagen
Collagen () is the main structural protein in the extracellular matrix of the connective tissues of many animals. It is the most abundant protein in mammals, making up 25% to 35% of protein content. Amino acids are bound together to form a trip ...
gene can lead to the disease Osteogenesis Imperfecta
Osteogenesis imperfecta (; OI), colloquially known as brittle bone disease, is a group of genetic disorders that all result in bones that bone fracture, break easily. The range of symptoms—on the skeleton as well as on the body's other Or ...
, which is characterized by brittle bones. In the illustration, a stem-loop formed from an imperfect inverted repeat is mutated with a thymine (T) nucleotide insertion as a result of an inter- or intrastrand switch. The addition of the T creates a base-pairing
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
"match up" with the adenine (A) that was previously a "bump" on the left side of the stem. While this addition makes the stem stronger and perfects the inverted repeat, it also creates a frameshift mutation
A frameshift mutation (also called a framing error or a reading frame shift) is a genetic mutation caused by indels ( insertions or deletions) of a number of nucleotides in a DNA sequence that is not divisible by three. Due to the triplet natur ...
in the nucleotide sequence which alters the reading frame
In molecular biology, a reading frame is a specific choice out of the possible ways to read the nucleic acid sequence, sequence of nucleotides in a nucleic acid (DNA or RNA) molecule as a sequence of triplets. Where these triplets equate to amino ...
and will result in an incorrect expression of the gene.
Programs and databases
The following list provides information and external links to various programs and databases for inverted repeats:non-B DB
A Database for Integrated Annotations and Analysis of non-B DNA Forming Motifs. This database is provided by The Advanced Biomedical Computing Center (ABCC) at then Frederick National Laboratory for Cancer Research (FNLCR). It covers the A-DNA and Z-DNA conformations otherwise known as "non-B DNAs" because they are not the more common B-DNA form of a right-handed Watson-Crick
double-helix
In molecular biology, the term double helix refers to the structure formed by double-stranded molecules of nucleic acids such as DNA. The double helical structure of a nucleic acid complex arises as a consequence of its secondary structure, a ...
. These "non-B DNAs" include left-handed Z-DNA, cruciform
A cruciform is a physical manifestation resembling a common cross or Christian cross. These include architectural shapes, biology, art, and design.
Cruciform architectural plan
Christian churches are commonly described as having a cruciform ...
, triplex, tetraplex and hairpin structures. Searches can be performed on a variety of "repeat types" (including inverted repeats) and on several species.Inverted Repeats Database
Boston University. This database is a web application that allows query and analysis of repeats held in the PUBLIC DATABASE project. Scientists can also analyze their own sequences with the Inverted Repeats Finder algorithm.
P-MITE: a Plant MITE database
— this database for
Miniature Inverted-repeat Transposable Elements Miniature Inverted-repeat Transposable Elements (MITEs) are a group of non-autonomous Class II transposable elements (DNA sequences). Being non-autonomous, MITEs cannot code for their own transposase. They exist within the genomes of animals, plant ...
(MITEs) contains sequences from plant genomes. Sequences may be searched or downloaded from the database.
* EMBOSS is the "European Molecular Biology Open Software Suite" which runs on UNIX
Unix (, ; trademarked as UNIX) is a family of multitasking, multi-user computer operating systems that derive from the original AT&T Unix, whose development started in 1969 at the Bell Labs research center by Ken Thompson, Dennis Ritchie, a ...
and UNIX-like
A Unix-like (sometimes referred to as UN*X, *nix or *NIX) operating system is one that behaves in a manner similar to a Unix system, although not necessarily conforming to or being certified to any version of the Single UNIX Specification. A Uni ...
operating systems. Documentation and program source files are available on thEMBOSS website
Applications specifically related to inverted repeats are listed below:
Finds inverted repeats in nucleotide sequences. Threshold values can be set to limit the scope of the search.
EMBOSS palindrome
Finds
palindromes
A palindrome ( /ˈpæl.ɪn.droʊm/) is a word, number, phrase, or other sequence of symbols that reads the same backwards as forwards, such as ''madam'' or '' racecar'', the date " 02/02/2020" and the sentence: "A man, a plan, a canal – Pana ...
such as stem loop regions in nucleotide sequences. The program will find sequences that include sections of mismatches and gaps that may correspond to bulges in a stem loop.
References
External links
* {{Repeated sequence Repetitive DNA sequences Molecular biology