Formylglycine-generating Enzyme on:
[Wikipedia]
[Google]
[Amazon]
Formylglycine-generating enzyme (FGE), located at 3p26.1 in humans, is the name for an
enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. A ...
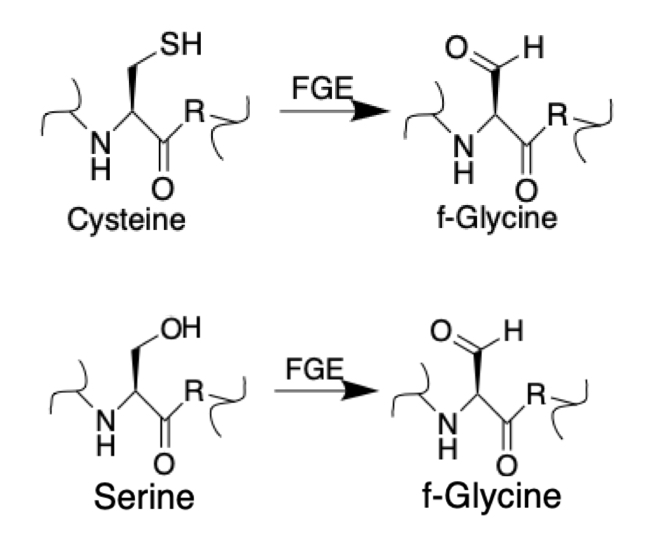
present in the endoplasmic reticulum that catalyzes the conversion of cysteine
Cysteine (symbol Cys or C; ) is a semiessential proteinogenic amino acid with the formula . The thiol side chain in cysteine often participates in enzymatic reactions as a nucleophile.
When present as a deprotonated catalytic residue, sometime ...
to formylglycine (fGly). There are two main classes of FGE, aerobic and anaerobic. FGE activates sulfatases, which are essential for the degradation of sulfate ester
Organosulfates are a class of organic compounds sharing a common functional group with the structure R-O-SO3−. The SO4 core is a sulfate group and the R group is any organic residue. All organosulfates are formally esters derived from alcohols ...
s. The catalytic activity of sulfatases is dependent upon a formylglycine (sometimes called oxoalanine) residue in the active site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate (binding site) a ...
.
Aerobic
The aerobic enzyme has astructure
A structure is an arrangement and organization of interrelated elements in a material object or system, or the object or system so organized. Material structures include man-made objects such as buildings and machines and natural objects such as ...
homologous to the complex alpha/beta topology found in the gene product of human sulfatase-modifying factor 1 (SUMF1
Sulfatase-modifying factor 1 is an enzyme that in humans is encoded by the ''SUMF1'' gene.
Sulfatases catalyze the hydrolysis of sulfate esters such as glycosaminoglycans, sulfolipids, and steroid sulfates. C-alpha-formylglycine (FGly), the catal ...
). Aerobic FGE converts a cysteine residue in the highly conserved consensus sequence CXPXR to fGly. To do so, FGE “activates” its target by utilizing mononuclear copper. The substrate first binds to copper, increasing reactivity of the substrate-copper complex with oxygen. Activation is then accomplished through oxidation of a cysteine residue in the substrate-copper complex. Due to the nature of this reaction, FGE is termed a “copper-dependent metalloenzyme. 
Anaerobic
The most well-studied anaerobic FGE is the bacterial AtsB, an iron-sulfur cluster containing enzyme present in ''Klebsiella pneumoniae'', that is able to convert either cysteine or serine to fGly with a distinctly different mechanism than the aerobic form. While AtsB can convert either, its activity increases four fold when in the presence of cysteine over serine. AtsB is 48% similar to an enzyme present in ''Clostridium perfringens''. Both enzymes possess the Cx3Cx2C motif unique to the radical S-adenosyl methionine superfamily and are able to use a reduction reaction to cleave S-adenosyl methionine. These two enzymes fall into a larger group called the anaerobic Sulfatase Maturing Enzymes, which are able to convert cysteine into fGly without the use of oxygen.Protein domain
In molecular biology, "formylglycine-generating enzyme" (sometimes annotated as formylglycine-generating sulfatase enzyme) is the name of the FGE protein domain, whether or not the protein is catalytically active. Both prokaryotic and eukaryotic homologs of FGE possess highly conserved active sites — including the catalytic cysteine residues required for enzymatic function. Activation of molecular oxygen is thought to be carried out by conserved residues close to the FGE catalytic site in aerobic organisms. The catalytic cysteine residues are involved in a thiol-cysteine exchange leading to the ultimate production of fGly.Disease states
In humans, mutations in SUMF1 result in defects in FGE, which in turn causes the impairment of sulfatases. The result is a disease calledmultiple sulfatase deficiency
Multiple sulfatase deficiency (MSD), also known as Austin disease, or mucosulfatidosis, is a very rare autosomal recessiveJames, William; Berger, Timothy; Elston, Dirk (2005). ''Andrews' Diseases of the Skin: Clinical Dermatology''. (10th ed.). Sau ...
(MSD), in which the accumulation of glycosaminoglycans
Glycosaminoglycans (GAGs) or mucopolysaccharides are long, linear polysaccharides consisting of repeating disaccharide units (i.e. two-sugar units). The repeating two-sugar unit consists of a uronic sugar and an amino sugar, except in the case ...
or sulfolipid
Sulfolipids are a class of lipids which possess a sulfur-containing functional group. An abundant sulfolipid is sulfoquinovosyl diacylglycerol, which is composed of a glycoside of sulfoquinovose and diacylglycerol. In plants, sulfoquinovosyl dia ...
s can cause early infant death. This disease can be further differentiated into neonatal, late infantile, and juvenile, with neonatal being the most severe. Common symptoms include ichthyosis, hypotonia, skeletal abnormalities, and overall cognitive decline. In 2017 Weidner et al., found an association with SUMF1 expression and chronic obstructive pulmonary disease (COPD) development. As of January 2020, there were more than 100 reported cases worldwide of MSD. Known substrates for SUMF1 are: N-acetylgalactosamine-6-sulfate sulfatase (GALNS
N-acetylgalactosamine-6-sulfatase is an enzyme that, in humans, is encoded by the ''GALNS'' gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''gener ...
), arylsulfatase A
Arylsulfatase A (or cerebroside-sulfatase) is an enzyme that breaks down sulfatides, namely cerebroside 3-sulfate into cerebroside and sulfate. In humans, arylsulfatase A is encoded by the ''ARSA'' gene.
Pathology
A deficiency is associated with m ...
(ARSA), steroid sulfatase
Steroid sulfatase (STS), or steryl-sulfatase (EC 3.1.6.2), formerly known as arylsulfatase C, is a sulfatase enzyme involved in the metabolism of steroids. It is encoded by the ''STS'' gene.
Reactions
This enzyme catalysis, catalyses the follow ...
(STS) and arylsulfatase E
Arylsulfatase E, also known as ARSE, is an enzyme that, in humans, is encoded by the ''ARSE'' gene.
Function
Arylsulfatase E is a member of the arylsulfatase subfamily of sulfatase enzymes that catalyze the hydrolysis of sulfate esters. It i ...
(ARSE); all molecules that contain cysteine. FGE converts this cysteine group into C-𝛼-formylglycine. SUMF1 occurs in the endoplasmic reticulum
The endoplasmic reticulum (ER) is, in essence, the transportation system of the eukaryotic cell, and has many other important functions such as protein folding. It is a type of organelle made up of two subunits – rough endoplasmic reticulum ( ...
or its lumen.
References
{{InterPro content, IPR005532 Protein domains