FANTOM on:
[Wikipedia]
[Google]
[Amazon]
FANTOM (Functional Annotation of the Mouse/Mammalian Genome) is an international research 
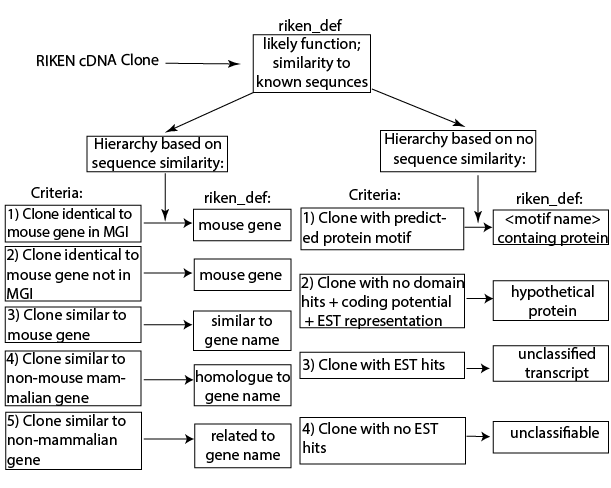 Central to the curation efforts was the creation of the RIKEN definition. This provided a
Central to the curation efforts was the creation of the RIKEN definition. This provided a


consortium
A consortium (plural: consortia) is an association of two or more individuals, companies, organizations or governments (or any combination of these entities) with the objective of participating in a common activity or pooling their resources for ...
first established in 2000 as part of the RIKEN research institute in Japan
Japan ( ja, 日本, or , and formally , ''Nihonkoku'') is an island country in East Asia. It is situated in the northwest Pacific Ocean, and is bordered on the west by the Sea of Japan, while extending from the Sea of Okhotsk in the north ...
. The original meeting gathered international scientists from diverse backgrounds to help annotate the function of mouse cDNA clones generated by the Hayashizaki group. Since the initial FANTOM1 effort, the consortium has released multiple projects that look to understand the mechanisms governing the regulation of mammalian genome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding ge ...
s. Their work has generated a large collection of shared data and helped advance biochemical
Biochemistry or biological chemistry is the study of chemical processes within and relating to living organisms. A sub-discipline of both chemistry and biology, biochemistry may be divided into three fields: structural biology, enzymology an ...
and bioinformatic
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combine ...
methodologies in genomics
Genomics is an interdisciplinary field of biology focusing on the structure, function, evolution, mapping, and editing of genomes. A genome is an organism's complete set of DNA, including all of its genes as well as its hierarchical, three-dim ...
research.

Foundation
In 1995, researchers of the RIKEN institute began creating anencyclopedia
An encyclopedia (American English) or encyclopædia (British English) is a reference work or compendium providing summaries of knowledge either general or special to a particular field or discipline. Encyclopedias are divided into articles ...
of full length cDNAs for the mouse
A mouse ( : mice) is a small rodent. Characteristically, mice are known to have a pointed snout, small rounded ears, a body-length scaly tail, and a high breeding rate. The best known mouse species is the common house mouse (''Mus musculus' ...
genome. The goal of this 'Mouse Encyclopedia Project' was to provide a functional annotation
An annotation is extra information associated with a particular point in a document or other piece of information. It can be a note that includes a comment or explanation. Annotations are sometimes presented in the margin of book pages. For anno ...
of the mouse transcriptome
The transcriptome is the set of all RNA transcripts, including coding and non-coding, in an individual or a population of cells. The term can also sometimes be used to refer to all RNAs, or just mRNA, depending on the particular experiment. The t ...
. This mapping would provide a valuable resource for gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a ba ...
discovery, understanding disease-causing genes and homology
Homology may refer to:
Sciences
Biology
*Homology (biology), any characteristic of biological organisms that is derived from a common ancestor
* Sequence homology, biological homology between DNA, RNA, or protein sequences
*Homologous chrom ...
across species
In biology, a species is the basic unit of classification and a taxonomic rank of an organism, as well as a unit of biodiversity. A species is often defined as the largest group of organisms in which any two individuals of the appropriate s ...
. This promised to be a formidable task from the onset. Current methodologies were insufficient to generate full length cDNA clones at scale, and to be useful as a resource the annotations would have to be agreed upon by experts across different disciplines.
The first goal was to develop methods that allowed generation of full length cDNA libraries. Reverse transcriptase
A reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, ...
protocols at the time had difficulties with the secondary structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta ...
of mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
, leading to abbreviated cDNAs that were difficult to align and invited further complications in downstream analysis. To surpass this limitation, a method utilizing trehalose
Trehalose (from Turkish '' tıgala'' – a sugar derived from insect cocoons + -ose) is a sugar consisting of two molecules of glucose. It is also known as mycose or tremalose. Some bacteria, fungi, plants and invertebrate animals synthesize it ...
was developed to allow reverse transcriptase to function at a higher temperature, relaxing secondary structures. Other methods were additionally developed to assist in the construction of clonal cDNA libraries. These include a biotin
Biotin (or vitamin B7) is one of the B vitamins. It is involved in a wide range of metabolic processes, both in humans and in other organisms, primarily related to the utilization of fats, carbohydrates, and amino acids. The name ''biotin'', bor ...
-based capture system to select for full length cDNA, a novel lambda phage
''Enterobacteria phage λ'' (lambda phage, coliphage λ, officially ''Escherichia virus Lambda'') is a bacterial virus, or bacteriophage, that infects the bacterial species ''Escherichia coli'' (''E. coli''). It was discovered by Esther Lederb ...
vector
Vector most often refers to:
*Euclidean vector, a quantity with a magnitude and a direction
*Vector (epidemiology), an agent that carries and transmits an infectious pathogen into another living organism
Vector may also refer to:
Mathematic ...
that minimized biases when delivering cDNA into a plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria; how ...
, and an iterative
Iteration is the repetition of a process in order to generate a (possibly unbounded) sequence of outcomes. Each repetition of the process is a single iteration, and the outcome of each iteration is then the starting point of the next iteration. ...
strategy to enrich for cDNA that had yet to be sequenced
In genetics and biochemistry, sequencing means to determine the primary structure (sometimes incorrectly called the primary sequence) of an unbranched biopolymer. Sequencing results in a symbolic linear depiction known as a sequence which suc ...
.
Sequencing began in 1998 and progressed rapidly, producing 246 cDNA libraries that encompassed 21,076 cDNA clones across a large range of mouse cells and tissues. While this stage was largely successful, further limitations were encountered at the bioinformatic level. The sequenced cDNAs were annotated in a semi-automatic manner that utilized available databases
In computing, a database is an organized collection of data stored and accessed electronically. Small databases can be stored on a file system, while large databases are hosted on computer clusters or cloud storage. The design of databases spa ...
(such as species homology and known protein motifs) to assign genes within a Gene Ontology
The Gene Ontology (GO) is a major bioinformatics initiative to unify the representation of gene and gene product attributes across all species. More specifically, the project aims to: 1) maintain and develop its controlled vocabulary of gene and g ...
(GO) framework. However, many novel sequences did not have meaningful matches when BLAST
Blast or The Blast may refer to:
*Explosion, a rapid increase in volume and release of energy in an extreme manner
*Detonation, an exothermic front accelerating through a medium that eventually drives a shock front
Film
* ''Blast'' (1997 film), ...
against gene databases.
After consulting Gerry Rubin
Gerald Mayer Rubin (born 1950) is an American biologist, notable for pioneering the use of transposable P elements in genetics, and for leading the public project to sequence the ''Drosophila melanogaster'' genome. Related to his genomics wor ...
, the organizer of the first genome annotation effort for Drosophila melanogaster
''Drosophila melanogaster'' is a species of fly (the taxonomic order Diptera) in the family Drosophilidae. The species is often referred to as the fruit fly or lesser fruit fly, or less commonly the "vinegar fly" or "pomace fly". Starting with Ch ...
, it became apparent that a robust system for annotation that incorporated computational prediction and manual curation was required for the novel sequences. Desiring input from experts in bioinformatics, genetics and other scientific fields, the RIKEN group organized the first FANTOM meeting.
FANTOM1
To facilitate the annotation of the mouse cDNA clones, the RIKEN research group developed a web-based service called FANTOM+ prior to the first meeting. Users could search for motifs, view pre-computed sequence similarity scores, as well as query other public databases and integrate relevant annotations into the FANTOM database. The assignment and functional annotation of the genes required multiple bioinformatic tools and databases. Predominant tools included BLASTN/BLASTX,FASTA
FASTA is a DNA and protein sequence alignment software package first described by David J. Lipman and William R. Pearson in 1985. Its legacy is the FASTA format which is now ubiquitous in bioinformatics.
History
The original FASTA program ...
/FASTY, DECODER, EST-WISE and HMMER
HMMER is a free and commonly used software package for sequence analysis written by Sean Eddy. Its general usage is to identify homologous protein or nucleotide sequences, and to perform sequence alignments. It detects homology by comparing ...
, while both nucleic acid
Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomers made of three components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main cl ...
and protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, respo ...
databases such as SwissProt, UniGene
UniGene was a NCBI database of the transcriptome and thus, despite the name, not primarily a database for genes. Each entry is a set of transcripts that appear to stem from the same transcription locus (i.e. gene or expressed pseudogene). Inform ...
and NCBI-nr were utilized. Concurrently, a collaboration with the Mouse Genome Informatics
Mouse Genome Informatics (MGI) is a free, online database and bioinformatics resource hosted by The Jackson Laboratory, with funding by the National Human Genome Research Institute (NHGRI), the National Cancer Institute (NCI), and the Eunice Kenne ...
group (MGI) allowed the RIKEN researchers to establish a validated set of clones that were identical between the two databases.
Armed with computational methodologies and over 20,000 cDNA sequences, the RIKEN group organized the first FANTOM meeting in Tsukuba City from August 28 to September 8, 2000. A diverse group of international scientists were recruited to discuss strategies and execute the annotation of the RIKEN clones. The assembled computational procedures allowed for sequence comparison and domain analysis to assign putative function using GO terms. Redundancy of the cDNA clones presented a challenge, requiring clustering strategies and referral to the MGI validation set to identify unique clones. The RIKEN set of clones was eventually reduced to 15,295 genes, although this was cautiously considered an overestimation.
 Central to the curation efforts was the creation of the RIKEN definition. This provided a
Central to the curation efforts was the creation of the RIKEN definition. This provided a hierarchical
A hierarchy (from Greek: , from , 'president of sacred rites') is an arrangement of items (objects, names, values, categories, etc.) that are represented as being "above", "below", or "at the same level as" one another. Hierarchy is an important ...
and systematic means to assign functions to the clones based upon known genes, placing priority on previously established or well-curated knowledge. The hierarchical nature of the classification Classification is a process related to categorization, the process in which ideas and objects are recognized, differentiated and understood.
Classification is the grouping of related facts into classes.
It may also refer to:
Business, organizat ...
allowed for consistency when a sequence was highly similar to multiple different genes. Importantly, if no sequence similarity was found, the definition assigned putative function based upon predicted protein motif signatures, coding potential and matches to expressed sequence tag In genetics, an expressed sequence tag (EST) is a short sub-sequence of a cDNA sequence. ESTs may be used to identify gene transcripts, and were instrumental in gene discovery and in gene-sequence determination. The identification of ESTs has proc ...
(EST) databases. Only in the absence of any predicted or representative similarity would a clone be considered ‘unclassifiable.’
The collected efforts of RIKEN/FANTOM resulted in a 2001 Nature publication. The results included the assignment of the 21,076 cDNA clones to 4,012 GO terms, identification of novel mouse genes and protein motifs, detection of likely alternative spliceforms, and the discovery of mouse genes orthologous
Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a spec ...
to human disease genes. Additionally, the first sequenced human genome was published a week later and incorporated FANTOM's results to predict the number of human genes.
FANTOM2
Having established and improved upon the protocols for full-length cDNA library generation, the RIKEN group continued to add to the FANTOM collection. Modifications to their methods allowed for further selection of rare and long transcripts, enabling identification of cDNA over 4kb in length. The second FANTOM meeting occurred May 2002 - by then the number of cDNA clones had increased by 39,694 to a total of 60,770. One insight gained from FANTOM1 was that alternativepolyadenylation
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In euk ...
was common in the mouse transcriptome, meaning that 3’-end clustering led to extensive redundancy. To address this, additional sequencing of the 5’-end was performed to identify unique clones. The FANTOM2 publication contributed a substantial addition of novel protein coding transcripts. Arguably the most notable result of FANTOM2 was that efforts to select for long and rare transcripts had revealed a significant amount of non protein-coding RNA.
Again, the FANTOM collection proved to be a fruitful resource. The non-coding RNA were identified as antisense RNA
Antisense RNA (asRNA), also referred to as antisense transcript, natural antisense transcript (NAT) or antisense oligonucleotide, is a single stranded RNA that is complementary to a protein coding messenger RNA (mRNA) with which it hybridizes, and ...
and long non-coding RNA
Long non-coding RNAs (long ncRNAs, lncRNA) are a type of RNA, generally defined as transcripts more than 200 nucleotides that are not translated into protein. This arbitrary limit distinguishes long ncRNAs from small non-coding RNAs, such as mic ...
s (lncRNA), poorly understood classes of regulatory RNA. The first published sequence of the mouse genome utilized the annotations established by FANTOM. Other efforts were able to describe entire protein families, such as the G protein-coupled receptor
G protein-coupled receptors (GPCRs), also known as seven-(pass)-transmembrane domain receptors, 7TM receptors, heptahelical receptors, serpentine receptors, and G protein-linked receptors (GPLR), form a large group of evolutionarily-related p ...
s.
FANTOM3
An ultimate goal of FANTOM is to establish gene networks that capture the regulatory interactions of transcription, and to differentiate these interactions bycell type
A cell type is a classification used to identify cells that share morphological or phenotypical features. A multicellular organism may contain cells of a number of widely differing and specialized cell types, such as muscle cells and skin cells, ...
or state. To this extent, it was realized that the polymorphic nature of the 5'-end of sequences would require extensive mapping. Characterizing transcription start sites (TSSs) would allow identification of promoters and differentiation of their usage between cell types. This also meant further developments in sequencing methods were needed. While full length mouse cDNAs continued to be generated, the RIKEN-led researchers established Cap Analysis of Gene Expression (CAGE), a technique that would drive much of their future work.

Development of CAGE
CAGE was a continuation of the concepts developed for FANTOM1 - and used extensively in the following projects - for capturing 5' mRNA caps. Unlike previous efforts to generate full length cDNA, CAGE examines fragments, or tags, that are 20–27 in length. This provided an economical and high-throughput means of mapping TSSs, including promoter structure and activity. The general steps are as follows: cDNA is reverse transcribed from mRNA using random or oligo dT primers. The cap trapper method is then employed to ensure selection of full length cDNA. This entails adding biotin to the 5' cap, and subsequent capture with streptavidin beads after anRNase
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within t ...
digestion step to remove single stranded RNA that has not hybridized to cDNA. Following cap trapping, the cDNA is separated from the RNA-cDNA hybrid. A double-stranded CAGE linker that is also biotinylated is ligated to the 5' end of the cDNA, and the second strand of the cDNA is synthesized. This resulting dual stranded DNA is digested with the Mme1 endonuclease
Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (without regard to sequence), while many, typically called restriction endonucleases ...
, cutting the CAGE linker and producing a 20-27bp CAGE tag. A second linker is added to the 3'-end and the tag is amplified by PCR. Finally, the CAGE tags are released from the 5' and 3' linkers. The tags can then be sequenced, concatenated or cloned. At the time, CAGE was carried out using the RISA 384 capillary sequencer that had been previously established by RIKEN.

Discoveries
The development of CAGE gave rise to a number of milestone findings. Importantly, RNA was found to be much more abundant in the mammalian transcriptome than previously thought, accompanied with the realization that the genome was pervasively transcribed. Combining the methods of CAGE, gene identification signatures, and gene signature cloning, the ‘transcriptional landscape’ of the mammalian genome was mapped, characterizing the pattern of transcription control signals and the transcripts they generate. It was discovered that there are many more transcripts than the estimated 22,000 genes in the mouse genome, and that many of these transcriptional units have alternative promoters andpolyadenylation
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In euk ...
sites.
Furthermore, it was discovered that ‘transcriptional forests’, clusters of transcripts that share common expression regions and regulatory events, are separated by ‘transcription deserts,’ and make up ~63% of the genome. A jointly released publication found that many of the transcripts in these forests show antisense
In molecular biology and genetics, the sense of a nucleic acid molecule, particularly of a strand of DNA or RNA, refers to the nature of the roles of the strand and its complement in specifying a sequence of amino acids. Depending on the context, ...
transcription, and that most sense/antisense pairs show concordant regulation. Another notable result showed that many non-coding RNAs are dynamically expressed, with many being initiated in 3’ untranslated regions
In molecular genetics, an untranslated region (or UTR) refers to either of two sections, one on each side of a coding sequence on a strand of mRNA. If it is found on the 5' side, it is called the 5' UTR (or leader sequence), or if it is foun ...
, and that they are positionally conserved across species.
The third milestone paper to come out of FANTOM3 investigated mammalian promoter architecture and evolution. It established two classes of mammalian promoters. The first are TATA box
In molecular biology, the TATA box (also called the Goldberg–Hogness box) is a sequence of DNA found in the core promoter region of genes in archaea and eukaryotes. The bacterial homolog of the TATA box is called the Pribnow box which has ...
-enriched promoters, with well defined transcriptional start sites. These promoters are evolutionary conserved and are more commonly associated with tissue-specific genes. The second and more common class of promoters, broad CpG rich promoters, are plastic, evolvable, and expressed in a wide range of cells and tissues. This study also demonstrated that CpG-rich promoters may be bidirectional (produce sense-antisense pairs), and are highly susceptible epigenetic
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are "o ...
control and are thus a potential component of adaptive evolution
In biology, adaptation has three related meanings. Firstly, it is the dynamic evolutionary process of natural selection that fits organisms to their environment, enhancing their evolutionary fitness. Secondly, it is a state reached by the po ...
.
The meeting for FANTOM3 occurred in September, 2004. A collection of satellite publications that spawned from FANTOM3 were published in PLoS Genetics
''PLOS Genetics'' is a peer-reviewed open access scientific journal established in 2005 and published by the Public Library of Science. The founding editor-in-chief was Wayne N. Frankel (Columbia University Medical Center). The current editors-in ...
. They include further work on promoter properties, exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequen ...
length and pseudo-messenger RNA.
FANTOM4
The rise ofnext-generation sequencing Massive parallel sequencing or massively parallel sequencing is any of several high-throughput approaches to DNA sequencing using the concept of massively parallel processing; it is also called next-generation sequencing (NGS) or second-generation s ...
was significantly beneficial to the advancement of CAGE technology. Using the Roche-454 sequencer, the FANTOM group developed deepCAGE, increasing the throughput of CAGE to more than a million tags per sample. At these depths, researchers could now start constructing networks of gene regulatory interactions. The FANTOM4 meeting took place December, 2006.
While previous FANTOM projects examined a range of cell types, FANTOM4's purpose was to deeply interrogate the dynamics driving cellular differentiation
Cellular differentiation is the process in which a stem cell alters from one type to a differentiated one. Usually, the cell changes to a more specialized type. Differentiation happens multiple times during the development of a multicellular ...
. Analysis was confined to a human THP-1 cell line
THP-1 is a human monocytic cell line derived from an acute monocytic leukemia patient. It is used to test leukemia cell lines in immunocytochemical analysis of protein-protein interactions, and immunohistochemistry.
Characteristics
Although ...
, providing time course data of a monoblast
Monoblasts are the committed progenitor cells that differentiated from a committed macrophage or dendritic cell precursor (MDP) in the process of hematopoiesis. They are the first developmental stage in the monocyte series leading to a macrophage ...
becoming a monocyte
Monocytes are a type of leukocyte or white blood cell. They are the largest type of leukocyte in blood and can differentiate into macrophages and conventional dendritic cells. As a part of the vertebrate innate immune system monocytes also inf ...
. DeepCage resolved TSSs at single-nucleotide resolution, pinpointing where transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The fu ...
s (TFs) bind. By monitoring time-dependent gene expression changes as cells differentiated, inference was provided for which regulatory motifs are predictive of expression changes, time dependency of TF activity, and TF target genes. These efforts resulted in a transcriptional regulatory network, demonstrating that the differentiation process is highly complex and driven by a great magnitude of TFs enacting both positive and negative regulatory interactions.
FANTOM4 also increased our understanding of retrotransposon
Retrotransposons (also called Class I transposable elements or transposons via RNA intermediates) are a type of genetic component that copy and paste themselves into different genomic locations (transposon) by converting RNA back into DNA through ...
transcription and transcriptional initiation RNAs (tiRNAs). Retrotransposons contribute to repetitive elements in mammalian genomes and can affect multiple biological processes - like genomic evolution - as well as structures, such as alternative promoters and exons. It was demonstrated that retrotransposons are expressed in a cell and tissue specific manner, and approximately 250,000 previously unknown retrotransposon-driven TSSs were identified.
It was discovered that retrotransposons can influence mammalian transcription and transcriptional regulation of both coding and non-coding RNAs in various tissues. Further efforts found
a genomically and evolutionary widespread new class of RNAs, called transcription initiation RNAs (tiRNA). This species of RNA are relatively tiny (~18 nucleotides long) and are typically found downstream of TSSs of CpG rich promoters. tiRNAs are low in abundance and are associated with highly expressed genes, as well as RNA polymerase II
RNA polymerase II (RNAP II and Pol II) is a multiprotein complex that transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNAP enzymes found in the nucleus of eukaryoti ...
binding and TSSs. More recent work has shown that tiRNs may be able to modulate epigenetic states and local chromatin architecture. However, it possible that these tiRNAs do not have a regulatory role and are simply a byproduct of transcription.
Following these initial findings, an atlas of combinatorial transcriptional regulation in mouse and humans was published by the RIKEN researchers. This work demonstrated that transcriptional complexes can interact within a network to control tissue identity/cell state, and that these networks are often dominated by ‘facilitator' transcription factors which are broadly expressed across tissues/cells. It was found that about half of the measured regulatory interactions were conserved between mouse and human. FANTOM4 led to numerous satellite papers, investigating topics like promoter architecture, miRNA regulation and genomic regulatory blocks.
FANTOM5
The fifth round of FANTOM aimed to provide insight into the regulatory landscape of the transcriptome across as many cell states as possible. It continues to be a relevant resource of shared data. The project consisted of two phases: the first focused on steady state cells, while the second focused on temporal data. Advancements in next generation sequencing were leveraged to achieve FANTOM5’s great breadth, with single molecule sequencing allowing single base pair resolution of TSS activity from as little as 100 ng of RNA. Samples was collected from every human organ, as well as over 200cancer
Cancer is a group of diseases involving abnormal cell growth with the potential to invade or spread to other parts of the body. These contrast with benign tumors, which do not spread. Possible signs and symptoms include a lump, abnormal b ...
lines, 30 time courses of cellular differentiation, mouse development time courses, and over 200 primary cell types. In total, 1,816 human and 1,1016 mouse samples were profiled across both phases.
While similar to the ENCODE
The Encyclopedia of DNA Elements (ENCODE) is a public research project which aims to identify functional elements in the human genome.
ENCODE also supports further biomedical research by "generating community resources of genomics data, software ...
Project, FANTOM5 differs in two key ways. First, ENCODE utilized immortalised cell lines, while FANTOM5 focused on primary cells and tissues, which are more reflective of the actual biological processes responsible for maintaining cell type identity. Second, ENCODE utilized multiple genomic assays to capture the transcriptome and epigenome
An epigenome consists of a record of the chemical changes to the DNA and histone proteins of an organism; these changes can be passed down to an organism's offspring via transgenerational stranded epigenetic inheritance. Changes to the epigenome ...
. FANTOM5 focused solely on the transcriptome, relying on other published work to infer features like cell type as defined by chromatin status. The FANTOM5 meeting took place October, 2011.
Phase 1
The first phase of FANTOM5 involved taking ‘snapshots’ of a wide range of steady state cell types using CAGE profiling across 975 human and 399 mouse samples. This initial effort resulted in two Nature papers - one describing the mammalian promoter landscape and the other describing activeenhancers
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcriptio ...
. Together, they provide an atlas of promoters, enhancers and TSSs across diverse cell types, acting as a ‘baseline’ for studying the complex landscape of transcription regulation. Specifically, single molecule CAGE profiles were generated using a HeliScope sequencer across 573 human primary cell samples, 128 mouse primary cell samples, 250 cancer cell lines, 152 human post-mortem tissues and 271 mouse developmental tissue samples.
A new method to identify the CAGE peaks was developed, called decomposition peak analysis. CAGE tags are clustered by proximity, followed by independent component analysis
In signal processing, independent component analysis (ICA) is a computational method for separating a multivariate signal into additive subcomponents. This is done by assuming that at most one subcomponent is Gaussian and that the subcomponents ar ...
to decompose the peaks into non-overlapping regions. An enrichment step is applied to ensure the peaks correspond to TSSs, and external data of EST, histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn a ...
H3 lysine 4 trimethylation marks and DNase hypersensitivity sites are used to support that the peaks are genuine TSSs.
A key finding showed that the typical mammalian promoter contains multiple TSSs with differing expression patterns across samples. This implied that these TSSs are regulated separately, despite being within close proximity. Ubiquitously expressed promoters had the highest conservation in their sequences, while cell-specific promoters were less conserved. A further prominent result suggested that enhancer-derived RNA (eRNA) are transcribed in a cell/tissue specific manner, reflective of the activity of that enhancer.
Phase 2
While the first phase was focused on a steady state representation of cell states, the second phase looked to explore the dynamic process of transitioning cell states through the use of time course data. Again, CAGE was employed - this time over 19 human and 14 mouse time courses covering a range of cell types and biological stimuli that represented 408 distinct time points. This included the differentiation ofstem cell
In multicellular organisms, stem cells are undifferentiated or partially differentiated cells that can differentiate into various types of cells and proliferate indefinitely to produce more of the same stem cell. They are the earliest type o ...
or committed progenitor cell
A progenitor cell is a Cell (biology), biological cell that can Cellular differentiation, differentiate into a specific cell type. Stem cells and progenitor cells have this ability in common. However, stem cells are less specified than progenitor ...
s towards their terminal fates, as well as fully differentiated cells responding to growth factors
A growth factor is a naturally occurring substance capable of stimulating cell proliferation, wound healing, and occasionally cellular differentiation. Usually it is a secreted protein or a steroid hormone. Growth factors are important for regu ...
or pathogens
In biology, a pathogen ( el, πάθος, "suffering", "passion" and , "producer of") in the oldest and broadest sense, is any organism or agent that can produce disease. A pathogen may also be referred to as an infectious agent, or simply a germ ...
.
Unsupervised clustering was performed to identify a set of distinct response classes, examining patterns in expression fold changes compared to time 0. In this manner, the expression of enhancers, TF promoters and non-TF promoters were generalized on a temporal scale of the first 6 hours of the time-course. Generally, the earliest response of the cells occurred at enhancers, with eRNA concentrations peaking as early as 15 minutes after time 0. Even in the classes that represent ‘later’ responses, enhancers tended to activate before proximal promoters. Variability was seen in the persistence of this activation - some enhancers rapidly returned to baseline after the burst at 15 minutes, while others persisted after promoter activation. Together, this is suggestive that eRNA may have differential roles in regulating gene activity.
Additional Work
Aside from the typical sharing of data on the FANTOM database, FANTOM5 also introduced two bioinformatic tools for data exploration. ZENBU is a genome browser with additional functionality: users can upload BAM files of CAGE, short-RNA andChIP-seq
ChIP-sequencing, also known as ChIP-seq, is a method used to analyze protein interactions with DNA. ChIP-seq combines chromatin immunoprecipitation (ChIP) with massively parallel DNA sequencing to identify the binding sites of DNA-associated prote ...
experiments and perform quality control, normalization, peak finding and annotation among visual comparisons. SSTAR (Semantic catalog of, samples, transcription initiations, and regulations) meanwhile allows exploration and searches of the FANTOM5 samples and their genomic features.
The bounty of data produced by FANTOM5 continues to provide a resource for researchers looking to explain the regulatory mechanisms that shape processes like development. Often CAGE data in a specific cell/tissue type is used in conjunction with further epigenomic assays - one such example describes the interplay of DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts t ...
and CAGE-defined regulatory sequences during differentiation of a granulocyte
Granulocytes are
cells in the innate immune system characterized by the presence of specific granules in their cytoplasm. Such granules distinguish them from the various agranulocytes. All myeloblastic granulocytes are polymorphonuclear. They ha ...
.
Three years after introducing the enhancer and promoter atlases, the FANTOM group released atlases for lncRNAs and microRNA
MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides. Found in plants, animals and some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRN ...
s (miRNA), incorporating FANTOM5 data. An overarching goal was to provide further insight into the earlier observation of pervasive transcription of the mammalian genome. The lncRNA work characterized 27,919 human lncRNA genes across 1,829 samples to stimulate research in the functional relevance of this poorly understood class of RNA. The results were suggestive that 69% of the identified lncRNA had potential functionality, although more evidence is required to comment on whether the remaining 31% are merely transcriptional ‘noise’ from spurious transcription initiation. The miRNA atlas identified 1,357 human and 804 mouse miRNA promoters and demonstrated strong sequence conservation between the two species. It was also demonstrated that primary miRNA expression could be used as a proxy for mature miRNA levels.
FANTOM6
Currently underway, FANTOM6 aims to systematically characterize the role of lncRNA in the human genome. The biological function of these large (200+ nucleotides) and untranslated RNA is largely unknown. Based upon the few works that have examined lncRNA, it is believed that they are involved in regulating transcription,translation
Translation is the communication of the Meaning (linguistic), meaning of a #Source and target languages, source-language text by means of an Dynamic and formal equivalence, equivalent #Source and target languages, target-language text. The ...
, post-translational modifications, and epigenetic marks. However, current knowledge of the extent and range of these putative regulatory interactions is rudimentary.
There are numerous challenges to address for this next rendition of FANTOM. In particular, lncRNAs are ill-defined - they lack conservation and vary greatly in size, ranging from 200 to over one million nucleotides
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules w ...
in length. Unlike coding transcripts, which are found in the cytosol
The cytosol, also known as cytoplasmic matrix or groundplasm, is one of the liquids found inside cells (intracellular fluid (ICF)). It is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondri ...
for translation, lncRNA are found primarily in the nucleus
Nucleus ( : nuclei) is a Latin word for the seed inside a fruit. It most often refers to:
*Atomic nucleus, the very dense central region of an atom
*Cell nucleus, a central organelle of a eukaryotic cell, containing most of the cell's DNA
Nucle ...
- a much more complex landscape of RNA. In general, lncRNA have lower expression levels than coding transcripts, but there is great variability in this expression which can be obscured by cell type or localization within the nucleus. Furthermore, functional classification lncRNAs remains hotly debated - it is unknown if lncRNAs can be grouped based on common function/mechanisms of action, or by active domains.
FANTOM has laid out a three pronged experimental strategy to explore these unknowns. A reference transcriptome and epigenome profile of different cell types will be constructed as a base line for each cell type. Next, using lncRNAs identified in previous publications, FANTOM5 data and further CAGE profiling, perturbation experiments will be conducted to evaluate changes in cellular molecular phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology or physical form and structure, its developmental processes, its biochemical and physiological proper ...
. Lastly, complementary technology will be used to functionally annotate/classify a selected subset of lncRNAs. These techniques will be aimed at elucidating lncRNA secondary structure, their association to proteins and chromatin, and mapping long range interactions of lncRNA throughout the genome.
References
{{reflist Biological databases