|
RNase
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 (for the phosphorolytic enzymes) and 3.1 (for the hydrolytic enzymes) classes of enzymes. Function All organisms studied contain many RNases of two different classes, showing that RNA degradation is a very ancient and important process. As well as clearing of cellular RNA that is no longer required, RNases play key roles in the maturation of all RNA molecules, both messenger RNAs that carry genetic material for making proteins and non-coding RNAs that function in varied cellular processes. In addition, active RNA degradation systems are the first defense against RNA viruses and provide the underlying machinery for more advanced cellular immune strategies such as RNAi. Some cells also secrete copious quantities of non-specific RN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
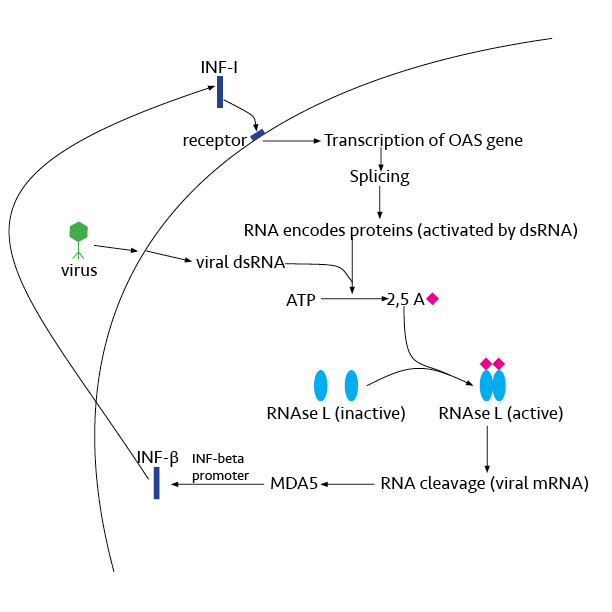
RNase L
Ribonuclease L or RNase L (for ''latent''), known sometimes as ribonuclease 4 or 2'-5' oligoadenylate synthetase-dependent ribonuclease — is an interferon (IFN)-induced ribonuclease which, upon activation, destroys all RNA within the cell (both cellular and viral). RNase L is an enzyme that in humans is encoded by the ''RNASEL'' gene. This gene encodes a component of the interferon-regulated 2'-5'oligoadenylate (2'-5'A) system that functions in the antiviral and antiproliferative roles of interferons. RNase L is activated by dimerization, which occurs upon 2'-5'A binding, and results in cleavage of all RNA in the cell. This can lead to activation of MDA5, an RNA helicase involved in the production of interferons. Synthesis and activation RNase L is present in very minute quantities during the normal cell cycle. When interferon binds to cell receptors, it activates transcription of around 300 genes to bring about the antiviral state. Among the enzymes produced is RNase L, w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Self-incompatibility
Self-incompatibility (SI) is a general name for several genetic mechanisms that prevent self-fertilization in sexually reproducing organisms, and thus encourage outcrossing and allogamy. It is contrasted with separation of sexes among individuals (dioecy), and their various modes of spatial (herkogamy) and temporally (dichogamy) separation. SI is best-studied and particularly common in flowering plants, although it is present in other groups, including sea squirts and fungi. In plants with SI, when a pollen grain produced in a plant reaches a stigma of the same plant or another plant with a matching allele or genotype, the process of pollen germination, pollen-tube growth, ovule fertilization, or embryo development is inhibited, and consequently no seeds are produced. SI is one of the most important means of preventing inbreeding and promoting the generation of new genotypes in plants and it is considered one of the causes of the spread and success of angiosperms on Earth. Mechan ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase T1
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 (for the phosphorolytic enzymes) and 3.1 (for the hydrolytic enzymes) classes of enzymes. Function All organisms studied contain many RNases of two different classes, showing that RNA degradation is a very ancient and important process. As well as clearing of cellular RNA that is no longer required, RNases play key roles in the maturation of all RNA molecules, both messenger RNAs that carry genetic material for making proteins and non-coding RNAs that function in varied cellular processes. In addition, active RNA degradation systems are the first defense against RNA viruses and provide the underlying machinery for more advanced cellular immune strategies such as RNAi. Some cells also secrete copious quantities of non-specific RN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase A
Pancreatic ribonuclease family (, ''RNase'', ''RNase I'', ''RNase A'', ''pancreatic RNase'', ''ribonuclease I'', ''endoribonuclease I'', ''ribonucleic phosphatase'', ''alkaline ribonuclease'', ''ribonuclease'', ''gene S glycoproteins'', ''Ceratitis capitata alkaline ribonuclease'', ''SLSG glycoproteins'', ''gene S locus-specific glycoproteins'', ''S-genotype-assocd. glycoproteins'', ''ribonucleate 3'-pyrimidino-oligonucleotidohydrolase'') is a superfamily of pyrimidine-specific endonucleases found in high quantity in the pancreas of certain mammals and of some reptiles. Specifically, the enzymes are involved in endonucleolytic cleavage of 3'-phosphomononucleotides and 3'-phosphooligonucleotides ending in C-P or U-P with 2',3'-cyclic phosphate intermediates. Ribonuclease can unwind the RNA helix by complexing with single-stranded RNA; the complex arises by an extended multi-site cation-anion interaction between lysine and arginine residues of the enzyme and phosphate groups of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase PhyM
RNase PhyM is a type of endoribonuclease An endoribonuclease is a ribonuclease endonuclease. It cleaves either single-stranded or double-stranded RNA, depending on the enzyme. Example includes both single proteins such as RNase III, RNase A, RNase T1, RNase T2 and RNase H and also com ... which is sequence specific for single stranded RNAs. It cleaves 3'-end of unpaired A and U residues. External links * * Ribonucleases {{Hydrolase-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosome
Ribosomes ( ) are macromolecular machines, found within all cells, that perform biological protein synthesis (mRNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins (RPs or r-proteins). The ribosomes and associated molecules are also known as the ''translational apparatus''. Overview The sequence of DNA that encodes the sequence of the amino acids in a protein is transcribed into a messenger RNA chain. Ribosomes bind to messenger RNAs and use their sequences for determining the correct sequence of amino acids to generate a given protein. Amino acids are selected and carried to the ribosome by transfer RNA (tRNA) molecules, which enter the ribosome and bind to the messenger RNA chain via an anti-c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TRNA
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino acid sequence of proteins. tRNAs genes from Bacteria are typically shorter (mean = 77.6 bp) than tRNAs from Archaea (mean = 83.1 bp) and eukaryotes (mean = 84.7 bp). The mature tRNA follows an opposite pattern with tRNAs from Bacteria being usually longer (median = 77.6 nt) than tRNAs from Archaea (median = 76.8 nt), with eukaryotes exhibiting the shortest mature tRNAs (median = 74.5 nt). Transfer RNA (tRNA) does this by carrying an amino acid to the protein synthesizing machinery of a cell called the ribosome. Complementation of a 3-nucleotide codon in a messenger RNA (mRNA) by a 3-nucleotide anticodon of the tRNA results in protein synthesis based on the mRNA code. As such, tRNAs are a necessary component of translation, the biological ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribozyme
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demonstrated that RNA can be both genetic material (like DNA) and a biological catalysis, catalyst (like protein enzymes), and contributed to the RNA world hypothesis, which suggests that RNA may have been important in the evolution of prebiotic self-replicating systems. The most common activities of natural or in vitro-evolved ribozymes are the cleavage or ligation of RNA and DNA and peptide bond formation. For example, the smallest ribozyme known (GUGGC-3') can aminoacylate a GCCU-3' sequence in the presence of PheAMP. Within the ribosome, ribozymes function as part of the large subunit ribosomal RNA to link amino acids during Translation (biology), protein synthesis. They also participate in a variety of RNA processing reactions, including RNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase P
Ribonuclease P (, ''RNase P'') is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein-based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA molecules. Further, RNase P is one of two known multiple turnover ribozymes in nature (the other being the ribosome), the discovery of which earned Sidney Altman and Thomas Cech the Nobel Prize in Chemistry in 1989: in the 1970s, Altman discovered the existence of precursor tRNA with flanking sequences and was the first to characterize RNase P and its activity in processing of the 5' leader sequence of precursor tRNA. Recent findings also reveal that RNase P has a new function. It has been shown that human nuclear RNase P is required for the normal and efficient transcription of various small noncoding RNAs, such as tRNA, 5S rRNA, SRP RNA and U6 snRNA genes, which a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme Commission Number
The Enzyme Commission number (EC number) is a numerical classification scheme for enzymes, based on the chemical reactions they catalyze. As a system of enzyme nomenclature, every EC number is associated with a recommended name for the corresponding enzyme-catalyzed reaction. EC numbers do not specify enzymes but enzyme-catalyzed reactions. If different enzymes (for instance from different organisms) catalyze the same reaction, then they receive the same EC number. Furthermore, through convergent evolution, completely different protein folds can catalyze an identical reaction (these are sometimes called non-homologous isofunctional enzymes) and therefore would be assigned the same EC number. By contrast, UniProt identifiers uniquely specify a protein by its amino acid sequence. Format of number Every enzyme code consists of the letters "EC" followed by four numbers separated by periods. Those numbers represent a progressively finer classification of the enzyme. Preliminary EC ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |